Biotype Determines Survival of Yersinia enterocolitica in Red Blood Cell Concentrates
- PMID: 40565236
- PMCID: PMC12193566
- DOI: 10.3390/ijms26125775
Biotype Determines Survival of Yersinia enterocolitica in Red Blood Cell Concentrates
Abstract
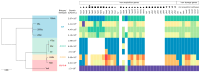
Red blood cell (RBC) concentrates remain at risk of bacterial contamination during cold storage. Although infrequent, Yersinia enterocolitica poses a significant blood safety risk. This study aimed to assess Y. enterocolitica bioserotype growth in RBC concentrates, serum sensitivity, and genetic diversity including iron metabolism genes. Ten Y. enterocolitica isolates from bioserotypes 1A, 1B/O:8, 4/O:3, and 2/O:9 were incubated in RBC concentrates and counted on days 3, 7, 14, 21, and 28. After incubation, the isolates were tested in human serum (NHS). Eight genomes were sequenced, analyzed using cgMLST, and screened for iron metabolism genes. The isolates formed two clusters, with 186dz (1A) and Ye8 (1B/O:8) as singletons. After 28 days in the RBC concentrates, the bacterial counts ranged from 1.98 × 10⁵ to 1.2 × 10⁹ CFU/mL, with Ye8 (1B/O:8) achieving the highest growth and one 4/O:3 isolate showing the lowest. All isolates survived 15-30 min in NHS, but the 28s isolate did not survive at 60 min. Serum sensitivity increased in two isolates, decreased in three, and remained unchanged in five. Isolates contained 27-42 iron metabolism genes with multiple allelic variants. The iron metabolism gene content or variants may influence the growth of Y. enterocolitica in RBC.
Keywords: Yersinia enterocolitica; bioserotypes; blood safety; contamination; red blood cells.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Hoelen D.W.M., Tjan D.H.T., Schouten M.A., Dujardin B.C.G., van Zanten A.R.H. Severe Yersinia Enterocolitica Sepsis after Blood Transfusion. Neth. J. Med. 2007;65:301–303. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

