DNA Satellites Impact Begomovirus Diseases in a Virus-Specific Manner
- PMID: 40565275
- PMCID: PMC12193555
- DOI: 10.3390/ijms26125814
DNA Satellites Impact Begomovirus Diseases in a Virus-Specific Manner
Abstract
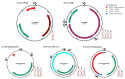
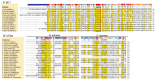
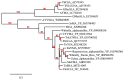
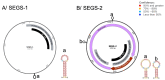
Begomoviruses infect many crops and weeds globally, especially in the tropical and subtropical regions, where there are waves of epidemics. These begomovirus epidemics are frequently associated with three DNA satellites: betasatellites, alphasatellites, and deltasatellites. Except for the origin of replication, these satellites show no sequence identity with the helper begomovirus. Alphasatellites and betasatellites encode the α-Rep and βC1 proteins, respectively, while deltasatellites encode no proteins. α-Rep, which functions like the Rep of the helper begomoviruses, ensures alphasatellite replication autonomy, while betasatellites and deltasatellites depend wholly on the helper virus for replication. The betasatellite βC1 protein is a pathogenicity determinant and suppressor of RNA silencing. The associations between satellites and helper viruses vary, depending on the virus and the host, and the roles of these satellites in disease development are an active area of investigation. This review highlights current information on the role of DNA satellites in begomovirus diseases and examines commonalities and differences between and within these satellites under prevailing conditions. Furthermore, two episomes, SEGS-1 and SEGS-2, associated with cassava mosaic geminiviruses, and their possible status as DNA satellites are discussed. DNA satellites are a major factor in begomovirus infections, which are a major constraint to crop production, especially in tropical and subtropical regions. Thus, areas for future research efforts, as well as implications in the biotechnological management of these viruses, are discussed in this review.
Keywords: alphasatellites; begomoviruses; betasatellites; deltasatellites.
Conflict of interest statement
The author declares no conflict of interest.
Figures




Similar articles
-
Differential interactions of ToLCNDV with different betasatellites reveal complex viral dynamics in N. benthamiana.PLoS One. 2025 Jun 25;20(6):e0327234. doi: 10.1371/journal.pone.0327234. eCollection 2025. PLoS One. 2025. PMID: 40561144 Free PMC article.
-
Molecular characterization of begomovirus and DNA satellites associated with mosaic and leaf curl disease of Jamaica cherry (Muntingia calabura) in India: Uncovering a new host for chilli leaf curl India virus.Virusdisease. 2024 Sep;35(3):484-495. doi: 10.1007/s13337-024-00891-w. Epub 2024 Sep 16. Virusdisease. 2024. PMID: 39464737
-
Advances in understanding begomovirus satellites.Annu Rev Phytopathol. 2013;51:357-81. doi: 10.1146/annurev-phyto-082712-102234. Annu Rev Phytopathol. 2013. PMID: 23915133 Review.
-
The Role of Satellites in the Evolution of Begomoviruses.Viruses. 2024 Jun 17;16(6):970. doi: 10.3390/v16060970. Viruses. 2024. PMID: 38932261 Free PMC article. Review.
-
Molecular diversity, recombination and population structure of alphasatellites associated with begomovirus disease complexes.Infect Genet Evol. 2017 Apr;49:39-47. doi: 10.1016/j.meegid.2017.01.001. Epub 2017 Jan 4. Infect Genet Evol. 2017. PMID: 28062387
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

