Exploring Carboxamide Derivatives as Promising Anticancer Agents: Design, In Vitro Evaluation, and Mechanistic Insights
- PMID: 40565364
- PMCID: PMC12193374
- DOI: 10.3390/ijms26125903
Exploring Carboxamide Derivatives as Promising Anticancer Agents: Design, In Vitro Evaluation, and Mechanistic Insights
Abstract
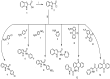
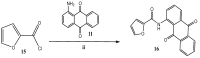
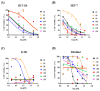
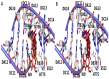
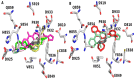
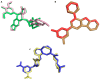
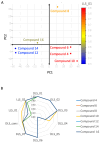
Carboxamide derivatives are a promising class of compounds in anticancer drug discovery, owing to their ability to interact with multiple oncogenic targets and their favorable pharmacological profiles. In this study, we report the design, synthesis, and biological evaluation of a series of N-substituted 1H-indole-2-carboxamides as potential anticancer agents. The synthesized compounds were assessed for antiproliferative activity using the MTT assay against MCF-7 (breast cancer), K-562 (leukemia), and HCT-116 (colon cancer) cell lines, with normal human dermal fibroblasts included as a non-cancerous control. Several compounds demonstrated notable cytotoxicity and selectivity. Compounds 12, 14, and 4 exhibited potent activity against K-562 cells, with IC50 values of 0.33 µM, 0.61 µM, and 0.61 µM, respectively. Compound 10 showed the most significant activity against HCT-116 cells (IC50 = 1.01 µM) with a high selectivity index (SI = 99.4). Moderate cytotoxicity was observed against MCF-7 cells. To elucidate the mechanism of action, molecular docking and induced-fit docking studies were conducted against key cancer-related targets, including topoisomerase-DNA (PDB ID: 5ZRF), PI3Kα (4L23), and EGFR (3W32), revealing favorable binding interactions. Additionally, principal component analysis of molecular descriptors indicated that the compounds possess promising drug-like and lead-like properties, particularly compound 10. Overall, this study highlights N-substituted indole-2-carboxamides as promising scaffolds for further optimization. The integration of synthetic chemistry, biological assays, and computational modeling provides a robust foundation for the continued development of these compounds as potential anticancer agents.
Keywords: MTT assay; carboxamides; cheminformatics; molecular docking; principal component analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Design, Synthesis, and Molecular Docking Studies of Indolo[3,2-c]Quinolines as Topoisomerase Inhibitors.Anticancer Agents Med Chem. 2025;25(14):1029-1040. doi: 10.2174/0118715206360700241219065917. Anticancer Agents Med Chem. 2025. PMID: 39902542
-
Design, synthesis, and biological evaluation of N-(2-amino-phenyl)-5-(4-aryl- pyrimidin-2-yl) amino)-1H-indole-2-carboxamide derivatives as novel inhibitors of CDK9 and class I HDACs for cancer treatment.Bioorg Chem. 2025 Jul 15;162:108577. doi: 10.1016/j.bioorg.2025.108577. Epub 2025 May 10. Bioorg Chem. 2025. PMID: 40383016
-
Design, synthesis, anti-breast cancer and in silico studies of substituted triphenyl imidazole-N-alkyl linked indole derivatives.Future Med Chem. 2025 Jul;17(14):1675-1692. doi: 10.1080/17568919.2025.2539673. Epub 2025 Aug 4. Future Med Chem. 2025. PMID: 40755380
-
Exploring the Structure-Activity Relationship of COX Inhibitors with Anticancer Effects: A Comprehensive Review.Curr Top Med Chem. 2025;25(9):1069-1104. doi: 10.2174/0115680266333495241011063253. Curr Top Med Chem. 2025. PMID: 39440774 Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Tarawneh N., Hamadneh L., Alshaer W., Al Bawab A.Q., Bustanji Y., Abdalla S. Downregulation of aquaporins and PI3K/AKT and upregulation of PTEN expression induced by the flavone scutellarein in human colon cancer cell lines. Heliyon. 2024;10:e39402. doi: 10.1016/j.heliyon.2024.e39402. - DOI - PMC - PubMed
-
- Cancer Facts & Figures. [(accessed on 14 September 2023)]. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

