Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer
- PMID: 40565366
- PMCID: PMC12192915
- DOI: 10.3390/ijms26125906
Comparison Bioinformatic Analysis of Extracellular Vesicles-Related Genes and MicroRNAs in Breast Cancer
Abstract
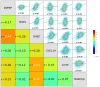
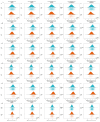
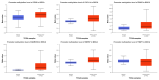
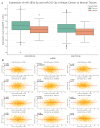
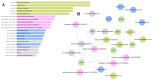
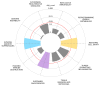
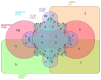
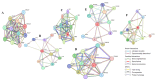
Breast cancer (BC) remains a leading cause of cancer-related mortality in women, with treatment challenges due to the lack of targeted therapies. Extracellular vesicles (EVs) play a crucial role in BC progression by carrying bioactive molecules. This study analyzed EV-associated molecules (ENPEP, TIMP1, CD36, MARCKS, DAB2, CXCL14, miR-181b-5p, miR-222-3p) using bioinformatics tools. We used GEPIA2; Human Protein Atlas (HPA) 24.0; bc-GenExMiner v5.1; UALCAN 2022; Kaplan-Meier plotter 2025; ENCORI database v2.0; Enrichr-KG web tool 2021; Cancer Hallmark Enrichment tool 2025; Tumor, Normal, and Metastatic (TNM) plot database 2025; MicroRNA Target Prediction Database 6.0; TargetScan 8.0; and STRING database 12.0. CD36, DAB2, and CXCL14 were significantly downregulated, while TIMP1 was upregulated in BC tissues (p < 0.05). CD36, CXCL14, and DAB2 were predominantly low in triple-negative and basal-like subtypes, whereas TIMP1 was higher in HER2+, ER+, and PR+ tumors (p < 0.01). These changes correlated with promoter methylation patterns. Higher TIMP1, DAB2, and CXCL14 levels were associated with improved overall survival (p < 0.05). miR-222-3p was downregulated and positively correlated with TIMP1 and DAB2, while miR-181b-5p was upregulated and negatively correlated with CXCL14. TNM analysis confirmed these expression changes. Functional enrichment linked these molecules to key cancer hallmarks, including proliferation and angiogenesis. CD36, DAB2, CXCL14, TIMP1, miR-222-3p, and miR-181b-5p may serve as biomarkers for BC pathogenesis and potential therapeutic targets. Further studies are needed to validate these findings.
Keywords: biomarkers; breast cancer; extracellular vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Lopez-Gonzalez L., Sanchez C.A., Sanchez C.C., Roberts C.E., Espinosa J., Pekarek T., Fraile-Martinez O., García-Montero C., Rodriguez-Slocker A.M., Jiménez-Álvarez L., et al. Exploring Biomarkers in Breast Cancer: Hallmarks of Diagnosis, Treatment, and Follow-Up in Clinical Practice. Medicina. 2024;60:168. doi: 10.3390/medicina60010168. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

