Cytokines from Macrophages Activated by Spike S1 of SARS-CoV-2 Cause eNOS/Arginase Imbalance in Endothelial Cells
- PMID: 40565379
- PMCID: PMC12192701
- DOI: 10.3390/ijms26125916
Cytokines from Macrophages Activated by Spike S1 of SARS-CoV-2 Cause eNOS/Arginase Imbalance in Endothelial Cells
Abstract
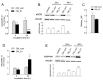
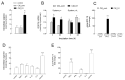
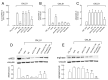
Multiple lines of evidence suggest that endothelial dysfunction is a key player in the pathogenesis of COVID-19, with cytokine storm as one of the main primary causes. Among the mechanisms underlying endothelial damage, clinical findings identify alterations in arginine metabolism, as patients with severe COVID-19 exhibit lower levels of nitric oxide synthase (eNOS) and upregulated arginase. In this study, we investigated, in human endothelial cells (HUVECs), the effect of conditioned medium from macrophages activated with SARS-CoV-2 Spike protein (CM_S1) on arginine metabolism. The results indicate that CM_S1 causes a marked decrease in eNOS and an increase in arginase, along with a greater intracellular arginine content and the induction of the CAT2 transporter. These effects are ascribable to the inflammatory mediators released by macrophages in CM_S1, mainly TNFα and IL-1β. Since infliximab, an antibody targeting TNFα, and baricitinib, an inhibitor of the JAK/STAT pathway, correct the observed imbalance between eNOS and arginase, our findings suggest the potential efficacy of a combined therapy to counteract endothelial dysfunction in COVID-19.
Keywords: TNFα; arginase; arginine; cytokines; eNOS; endothelial dysfunction; macrophages.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

