Comparative and Phylogenetic Analysis of the Complete Chloroplast Genomes of Lithocarpus Species (Fagaceae) in South China
- PMID: 40565508
- PMCID: PMC12192078
- DOI: 10.3390/genes16060616
Comparative and Phylogenetic Analysis of the Complete Chloroplast Genomes of Lithocarpus Species (Fagaceae) in South China
Abstract
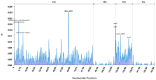
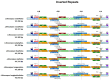
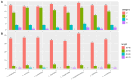
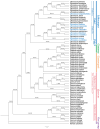
Background/Objectives: In South China, Lithocarpus species dominate mixed evergreen broadleaf forests, forming symbiotic relationships with ectomycorrhizal fungi and serving as food resources for diverse fauna, including frugivorous birds and mammals. The limited understanding of chloroplast genomes in this genus restricts our insights into its species diversity. This study investigates the chloroplast genome (cp genome) sequences from seven Lithocarpus species, aims to elucidate their structural variation, evolutionary relationships, and functional gene content to provide effective support for future genetic conservation and breeding efforts. Methods: We isolated total DNA from fresh leaves and sequenced the complete cp genomes of these samples. To develop a genomic resource and clarify the evolutionary relationships within Lithocarpus species, comparative chloroplast genome studies and phylogenetic investigations were performed. Results: All studied species exhibited a conserved quadripartite chloroplast genome structure, with sizes ranging from 161,495 to 163,880 bp. Genome annotation revealed 130 functional genes and a GC content of 36.72-37.76%. Codon usage analysis showed a predominance of leucine-encoding codons. Our analysis identified 322 simple sequence repeats (SSRs), which were predominantly palindromic in structure (82.3%). All eight species exhibited the same 19 SSR categories in similar proportions. Eight highly variable regions (ndhF, ycf1, trnS-trnG-exon1, trnk(exon1)-rps16(exon2), rps16(exon2), rbcL-accD, and ccsA-ndh) have been identified, which could be valuable as molecular markers in future studies on the population genetics and phylogeography of this genus. The phylogeny tree provided critical insights into the evolutionary trajectory of Fagaceae, suggesting that Lithocarpus was strongly supported as monophyletic, while Quercus was inferred to be polyphyletic, showing a significant cytonuclear discrepancy. Conclusions: We characterized and compared the chloroplast genome features across eight Lithocarpus species, followed by comprehensive phylogenetic analyses. These findings provide critical insights for resolving taxonomic uncertainties and advancing systematic research in this genus.
Keywords: Lithocarpus; chloroplast genomes; comparative genomics; genetic resources; phylogenetics analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- POWO Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. [(accessed on 9 April 2025)]. Available online: https://powo.science.kew.org/
-
- Webb C., Cannon C., Davies S. Tropical Forest Community Ecology. John Wiley & Sons; Hoboken, NJ, USA: 2008. Ecological organization, biogeography, and the phylogenetic structure of tropical forest tree communities; pp. 79–97.
-
- Mu X.Y., Li J.X., Xia X.F., Zhao L.C. Cupules and fruits of Lithocarpus (Fagaceae) from the Miocene of Yunnan, southwestern China. Taxon. 2015;64:795–808. doi: 10.12705/644.10. - DOI
-
- Cannon C.H., Manos P.S. Phylogeography of the Southeast Asian stone oaks (Lithocarpus) J. Biogeogr. 2003;30:211–226. doi: 10.1046/j.1365-2699.2003.00829.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

