Examination of Runs of Homozygosity Distribution Patterns and Relevant Candidate Genes of Potential Economic Interest in Russian Goat Breeds Using Whole-Genome Sequencing
- PMID: 40565522
- PMCID: PMC12192281
- DOI: 10.3390/genes16060631
Examination of Runs of Homozygosity Distribution Patterns and Relevant Candidate Genes of Potential Economic Interest in Russian Goat Breeds Using Whole-Genome Sequencing
Abstract
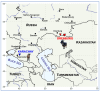
Background/objectives: Whole-genome sequencing (WGS) data provide valuable information about the genetic architecture of local livestock but have not yet been applied to Russian native goats, in particular, the Orenburg and Karachay breeds. A preliminary search for selection signatures based on single nucleotide polymorphism (SNP) genotype data in these breeds was not informative. Therefore, in this study, we aimed to address runs of homozygosity (ROHs) patterns and find the respective signatures of selection overlapping candidate genes in Orenburg and Karachay goats using the WGS approach.
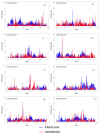
Methods: Paired-end libraries (150 bp reads) were constructed for each animal. Next-generation sequencing was performed using a NovaSeq 6000 sequencer (Illumina, Inc., San Diego, CA, USA), with ~20X genome coverage. ROHs were identified in sliding windows, and ROH segments shared by at least 50% of the samples were considered as ROH islands.
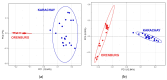
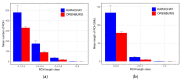
Results: ROH islands were identified on chromosomes CHI3, CHI5, CHI7, CHI12, CHI13, and CHI15 in Karachay goats; and CHI3, CHI11, CHI12, CHI15, and CHI16 in Orenburg goats. Shared ROH islands were found on CHI12 (containing the PARP4 and MPHOSPH8 candidate genes) and on CHI15 (harboring STIM1 and RRM1). The Karachay breed had greater ROH length and higher ROH number compared to the Orenburg breed (134.13 Mb and 695 vs. 78.43 Mb and 438, respectively). The genomic inbreeding coefficient (FROH) varied from 0.032 in the Orenburg breed to 0.054 in the Karachay breed. Candidate genes associated with reproduction, milk production, immunity-related traits, embryogenesis, growth, and development were identified in ROH islands in the studied breeds.
Conclusions: Here, we present the first attempt of elucidating the ROH landscape and signatures of selection in Russian local goat breeds using WGS analysis. Our findings will pave the way for further insights into the genetic mechanisms underlying adaption and economically important traits in native goats.
Keywords: Russian local breeds; candidate genes; goat (Capra hircus); runs of homozygosity (ROHs); signatures of selection; whole-genome sequencing (WGS).
Conflict of interest statement
The authors declare no conflicts of interest. The RSF had no role in the design of this study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Exploring Runs of Homozygosity and Heterozygosity in Sheep Breeds Maintained in Poland.Genes (Basel). 2025 Jun 14;16(6):709. doi: 10.3390/genes16060709. Genes (Basel). 2025. PMID: 40565601 Free PMC article.
-
Comparative Analysis of Runs of Homozygosity Islands in Indigenous and Commercial Chickens Revealed Candidate Loci for Disease Resistance and Production Traits.Vet Med Sci. 2025 Jan;11(1):e70074. doi: 10.1002/vms3.70074. Vet Med Sci. 2025. PMID: 39655377 Free PMC article.
-
Runs of homozygosity and selection signals analysis reveals domestication traits and divergence in local domestic duck breeds.Poult Sci. 2025 Jun 6;104(9):105404. doi: 10.1016/j.psj.2025.105404. Online ahead of print. Poult Sci. 2025. PMID: 40513394 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

