Satellite DNA Mapping in Suliformes (Aves): Insights into the Evolution of the Multiple Sex Chromosome System in Sula spp
- PMID: 40565525
- PMCID: PMC12193320
- DOI: 10.3390/genes16060633
Satellite DNA Mapping in Suliformes (Aves): Insights into the Evolution of the Multiple Sex Chromosome System in Sula spp
Abstract
Background: The order Suliformes exhibits significant karyotype diversity, with Sula species showing a Z1Z1Z2Z2/Z1Z2W multiple-sex chromosome system, an uncommon occurrence in avians. Satellite DNAs (satDNAs), which consist of tandemly repeated sequences, often vary considerably even among closely related species, making them valuable markers for studying karyotypic evolution, particularly that of sex chromosome evolution. This study aims to characterize and investigate the potential role of these sequences in the karyotypic evolution of the group, with special attention to the sex chromosomes.
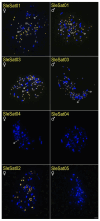
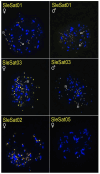
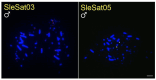
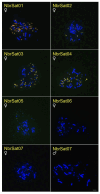
Methods: Through characterizing satDNAs in two Suliformes species (Sula leucogaster and Nannopterum brasilianum) using BGISEQ-500 platform and bioinformatics analysis. Their chromosomal distribution was mapped by fluorescence in situ hybridization (FISH) within their own karyotypes and in three additional Suliformes species (S. sula, S. dactylatra, and Fregata magnificens).
Results: Five satDNAs were identified in S. leucogaster and eight in N. brasilianum. Within the genus Sula, three species shared specific satDNA sequences, although with different hybridization patterns. In contrast, the satDNAs of N. brasilianum were species-specific. Additionally, the Z chromosome, including Z2 in Sula species, showed reduced accumulation of repetitive DNAs.
Conclusions: These results suggest that differential accumulation of repetitive sequences may have contributed to the diversification of karyotypes in this group, particularly influencing the structure and differentiation of sex chromosomes.
Keywords: chromosomal mapping; karyotypic evolution; multiple sex chromosomes; satellite DNA; suliformes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Karyotype evolution of suliformes and description of a ♂Z1Z1Z2Z2/♀Z1Z2W multiple sex chromosome system in boobies (Sula spp.).Genome. 2025 Jan 1;68:1-11. doi: 10.1139/gen-2024-0165. Genome. 2025. PMID: 39883916
-
New Insights into the Sex Chromosome Evolution of the Common Barker Frog Species Complex (Anura, Leptodactylidae) Inferred from Its Satellite DNA Content.Biomolecules. 2025 Jun 16;15(6):876. doi: 10.3390/biom15060876. Biomolecules. 2025. PMID: 40563516 Free PMC article.
-
Cytogenomic analysis in Seriemas (Cariamidae): Insights into an atypical avian karyotype.J Hered. 2025 Jul 21;116(4):441-452. doi: 10.1093/jhered/esaf012. J Hered. 2025. PMID: 40080045
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
References
-
- Schmidt T. Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends Plant Sci. 1998;3:195–199. doi: 10.1016/S1360-1385(98)01223-0. - DOI
-
- Plohl M., Meštrović N., Mravinac B. Satellite DNA Evolution. In: Garrido-Ramos M.A., editor. Repetitive DNA. Volume 7. S. Karger AG; Basel, Switzerland: 2012. pp. 126–152. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

