Comparative and Phylogenetic Analysis of Complete Chloroplast Genomes of Five Mangifera Species
- PMID: 40565558
- PMCID: PMC12192436
- DOI: 10.3390/genes16060666
Comparative and Phylogenetic Analysis of Complete Chloroplast Genomes of Five Mangifera Species
Abstract
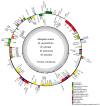
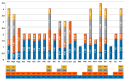
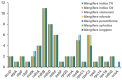
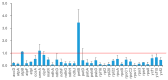
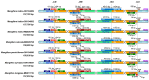
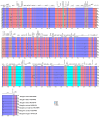
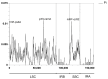
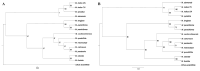
Background/Objectives: Mango, which is known as the "King of Tropical Fruits", is an evergreen plant belonging to the Anacardiaceae family. It belongs to the genus Mangifera, which comprises 69 species of plants found in tropical and subtropical regions, including India, Indonesia, the Malay Peninsula, Thailand, and South China. However, research on the structural information of complete chloroplast genomes of Mangifera is limited. Methods: The rapid advancement of high-throughput sequencing technology enables the acquisition of the entire chloroplast (cp) genome sequence, providing a molecular foundation for phylogenetic research. This work sequenced the chloroplast genomes of six Mangifera samples, performed a comparative analysis of the cp genomes, and investigated the evolutionary relationships within the Mangifera genus. Results: All six Mangifera samples showed a single circular molecule with a quadripartite structure, ranging from 157,604 bp to 158,889 bp in length. The number of RNA editing sites ranged from 60 to 61, with ndhB exhibiting the highest number of RNA editing sites across all species. Seven genes-namely, atpB, cemA, clpP, ndhD, petB, petD, and ycf15-exhibited a Ka/Ks value > 1, suggesting they may be under positive selection. Phylogenetic analysis revealed that Mangifera siamensis showed a close relationship between Mangifera indica and Mangifera sylvatica. Conclusions: Our comprehensive analysis of the whole cp genomes of the five Mangifera species offers significant insights regarding their phylogenetic reconstruction. Moreover, it elucidates the evolutionary processes of the cp genome within the Mangifera genus.
Keywords: Illumina; Mangifera; chloroplast genome; phylogenetic analysis; polymorphism analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Comparative and Phylogenetic Analysis of the Complete Chloroplast Genomes of Lithocarpus Species (Fagaceae) in South China.Genes (Basel). 2025 May 22;16(6):616. doi: 10.3390/genes16060616. Genes (Basel). 2025. PMID: 40565508 Free PMC article.
-
Three complete chloroplast genomes from two north American Rhus species and phylogenomics of Anacardiaceae.BMC Genom Data. 2024 Mar 15;25(1):30. doi: 10.1186/s12863-024-01200-6. BMC Genom Data. 2024. PMID: 38491489 Free PMC article.
-
Complete chloroplast genome of two Brachycorythis (Orchidaceae) species from China: comparative analysis and phylogenetic implications.BMC Genomics. 2025 Jul 2;26(1):632. doi: 10.1186/s12864-025-11791-8. BMC Genomics. 2025. PMID: 40604395 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Atypical antipsychotics for disruptive behaviour disorders in children and youths.Cochrane Database Syst Rev. 2017 Aug 9;8(8):CD008559. doi: 10.1002/14651858.CD008559.pub3. Cochrane Database Syst Rev. 2017. PMID: 28791693 Free PMC article.
Cited by
-
Comparative Genomics and Phylogenetics of Chloroplasts Reveal Lower Rates of Genetic Variation in Mango (Mangifera).Ecol Evol. 2025 Aug 8;15(8):e71957. doi: 10.1002/ece3.71957. eCollection 2025 Aug. Ecol Evol. 2025. PMID: 40785999 Free PMC article.
References
-
- Bajpai A., Muthukumar M., Ahmad I., Ravishankar K., Parthasarthy V., Sthapit B., Rao R., Verma J., Rajan S. Molecular and morphological diversity in locally grown non-commercial (heirloom) mango varieties of North India. J. Environ. Biol. 2016;37:221. - PubMed
-
- Iquebal M., Jaiswal S., Mahato A.K., Jayaswal P.K., Angadi U., Kumar N., Sharma N., Singh A.K., Srivastav M., Prakash J. MiSNPDb: A web-based genomic resources of tropical ecology fruit mango (Mangifera indica L.) for phylogeography and varietal differentiation. Sci. Rep. 2017;7:14968. doi: 10.1038/s41598-017-14998-2. - DOI - PMC - PubMed
-
- Dutta S., Srivastav M., Rymbai H., Chaudhary R., Singh A., Dubey A., Lal K. Pollen–pistil interaction studies in mango (Mangifera indica L.) cultivars. Sci. Hortic. 2013;160:213–221. doi: 10.1016/j.scienta.2013.05.012. - DOI
-
- Sherman A., Rubinstein M., Eshed R., Benita M., Ish-Shalom M., Sharabi-Schwager M., Rozen A., Saada D., Cohen Y., Ophir R. Mango (Mangifera indica L.) germplasm diversity based on single nucleotide polymorphisms derived from the transcriptome. BMC Plant Biol. 2015;15:277. doi: 10.1186/s12870-015-0663-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

