Natural Selection as the Primary Driver of Codon Usage Bias in the Mitochondrial Genomes of Three Medicago Species
- PMID: 40565565
- PMCID: PMC12193324
- DOI: 10.3390/genes16060673
Natural Selection as the Primary Driver of Codon Usage Bias in the Mitochondrial Genomes of Three Medicago Species
Abstract
Objectives: Codon usage bias is a fundamental feature of gene expression that can influence evolutionary processes and genetic diversity. This study aimed to investigate the mitochondrial codon usage characteristics and their driving forces in three Medicago species: Medicago polymorpha, Medicago sativa, and Medicago truncatula.
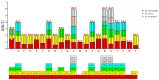
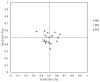
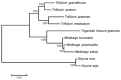
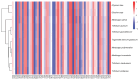
Methods: The complete mitochondrial genome sequences of the three species were downloaded from GenBank, and 21 shared coding sequences were screened. Codon usage patterns were analyzed using CodonW 1.4.2 and CUSP software. Key parameters, including the relative synonymous codon usage (RSCU), effective number of codons (ENC), codon adaptation index (CAI), codon bias index (CBI), and frequency of optimal codons (Fop), were calculated. Phylogenetic trees and RSCU clustering maps were constructed to explore evolutionary relationships.
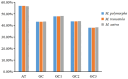
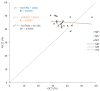
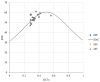
Results: The GC contents of the mitochondrial genomes followed the order of GC1 > GC2 > GC3. ENC values averaged above 35, while CAI, CBI, and Fop values ranged from 0.160 to 0.161, -0.078 to -0.076, and 0.362 to 0.363, respectively, indicating a weak preference for codons ending with A/U. Correlation and neutrality analyses suggested that codon usage bias was influenced by both mutation pressure and natural selection, with natural selection being the dominant factor. Fifteen optimal codons, predominantly ending with A/U, were identified. Phylogenetic analysis confirmed the close relationship among the three Medicago species, consistent with traditional taxonomy, whereas the RSCU clustering did not align with the phylogenetic relationships.
Conclusions: This study provides insights into the mitochondrial codon usage patterns and their evolutionary determinants in Medicago species, highlighting the predominant role of natural selection in shaping codon usage bias. The findings offer a foundation for comparative genomic studies and evolutionary analyses and may be beneficial for improving genetic engineering and breeding programs of Medicago species.
Keywords: Medicago; codon usage bias; mitochondrial genome; natural selection; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Codon usage characterization and phylogenetic analysis of the mitochondrial genome in Hemerocallis citrina.BMC Genom Data. 2024 Jan 13;25(1):6. doi: 10.1186/s12863-024-01191-4. BMC Genom Data. 2024. PMID: 38218810 Free PMC article.
-
Codon usage bias is presumably affected by tRNA selection effects in Actinidia polyploidization events.BMC Genomics. 2025 Jul 23;26(1):685. doi: 10.1186/s12864-025-11873-7. BMC Genomics. 2025. PMID: 40702430 Free PMC article.
-
Codon Usage Bias in Mitochondrial Genomes Across Three Species of Siphonaria (Mollusca: Gastropoda).Genes (Basel). 2025 Jun 26;16(7):747. doi: 10.3390/genes16070747. Genes (Basel). 2025. PMID: 40725404 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Individual-level interventions to reduce personal exposure to outdoor air pollution and their effects on people with long-term respiratory conditions.Cochrane Database Syst Rev. 2021 Aug 9;8(8):CD013441. doi: 10.1002/14651858.CD013441.pub2. Cochrane Database Syst Rev. 2021. PMID: 34368949 Free PMC article.
References
-
- Ma B., Liu C., Li Y., Wet Z., Cao D. Analysis and evaluation on the nutrition components of Huaiyang Medicago polymorpha with different mowing crops. Wei Sheng Yan Jiu J. Hyg. Res. 2015;44:532–537. - PubMed
-
- Al-Farsi S.M., Nawaz A., Anees-Ur-Rehman, Nadaf S.K., Al-Sadi A.M., Siddique K.H.M., Farooq M. Effects, tolerance mechanisms and management of salt stress in lucerne (Medicago sativa) Crop Pasture Sci. 2020;71:411–428. doi: 10.1071/CP20033. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

