Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia
- PMID: 40565569
- PMCID: PMC12193382
- DOI: 10.3390/genes16060677
Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia
Abstract
Objectives: Evaluate the genetic diversity and productive traits of crossbred cattle in the Caribbean region of Colombia, through analyses derived from the assessment of the genome-wide single-nucleotide polymorphism (SNP).
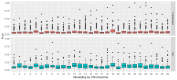
Methods: A total of 590 individuals and 66,098 SNPs were analyzed by principal components analysis (PCA) and detection of runs of homozygosity (ROH). The population was composed of 531 heifers marked as crossbreed and a group of 59 heifers marked as purebred Gyr. Additionally, allele frequencies were calculated for commercially important traits (CSN2, CSN3, LGB, DGAT1, GH1, CAPN1_316, CAPN1_350, CAPN1_4751, CAST_282, CAST_2870, and CAST_2959).
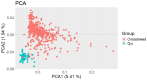
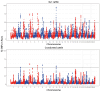
Results: Global differences in PCA were 7.35%, and principal components explained 1.94% and 5.41% of the variation. Five ROH islands were identified in crossbred animals on chromosomes 2, 5, 7, 8, and 12. The majority of observed ROH classes were shorter than 2 Mb, 54% in crossbreed cattle and 47% in Gyr cattle. Individual inbreeding was 5.2% in crossbreed and 12% in Gyr cattle. Both groups had similar allelic and genotypic frequencies for most of the evaluated commercial traits. Only a wide variation was observed in the genes related to growth hormone (GH1) and Calpastatin (CAST_2870 and CAST_22959). Crossbreed heifers had desired allele frequencies for better milk production and quality in the genes CSN2, LGB, DGAT1, and GH1, as well as in the genes CAST_2870 and CAST_2959.
Conclusions: Crossbreed cattle in the Colombian Caribbean region possess high genetic diversity and desirable allele frequencies to implement breeding and intense selection programs aimed at improving production yields.
Keywords: Gyr cattle; SNPs; dual-purpose systems; genomic analysis; homozygosity; inbreeding.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Differential A1/A2 β-casein (CSN2) gene-derived allelic and genotypic frequencies across Ecuadorian exotic dairy cattle breeds.Front Vet Sci. 2025 Jul 9;12:1616426. doi: 10.3389/fvets.2025.1616426. eCollection 2025. Front Vet Sci. 2025. PMID: 40703919 Free PMC article.
-
Exploring genetic diversity and genomic inbreeding across local beef cattle breeds.Animal. 2025 Jul;19(7):101565. doi: 10.1016/j.animal.2025.101565. Epub 2025 Jun 5. Animal. 2025. PMID: 40580843
-
Examination of Runs of Homozygosity Distribution Patterns and Relevant Candidate Genes of Potential Economic Interest in Russian Goat Breeds Using Whole-Genome Sequencing.Genes (Basel). 2025 May 24;16(6):631. doi: 10.3390/genes16060631. Genes (Basel). 2025. PMID: 40565522 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
References
-
- Brito L.F., Bédère N., Douhard F., Oliveira H.R., Arnal M., Peñagaricano F., Schinckel A.P., Baes C.F., Miglior F. Review: Genetic Selection of High-Yielding Dairy Cattle Toward Sustainable Farming Systems in a Rapidly Changing World. Animal. 2021;15:100292. doi: 10.1016/j.animal.2021.100292. - DOI - PubMed
-
- Herron J.C., Freeman S. Evolutionary Analysis. 5th ed. Pearson Education; London, UK: 2014. Mendelian Genetics in Populations II: Migration, Drift and Nonrandom Mating; pp. 259–264.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

