Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses
- PMID: 40565600
- PMCID: PMC12192454
- DOI: 10.3390/genes16060708
Half the Chromosome It Used to Be: Identifying Cancer Treatments Targeting Aneuploid Losses
Abstract
Background/objectives: Aneuploidy is near-ubiquitous in cancer and can decrease chemotherapy efficacy while also sensitizing cells to other drugs.
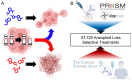
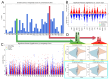
Methods: To systematically identify treatment strategies that target aneuploid cancers, data were integrated from The Cancer Genome Atlas (TCGA; 10,967 samples, 16,948 aneuploidy events) and the Broad Institute's Profiling Relative Inhibition Simultaneously in Mixtures (PRISM) screen of 578 cancer cell lines and 4518 compounds.
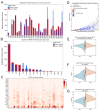
Results: Our analyses uncovered 37,720 significant positive and negative associations linking specific aneuploidies and treatments with patient prognosis or cell viability. Within TCGA data, 22 treatments correlated with improved 5-year survival for specific aneuploid cancers, whereas 46 were linked to worse outcomes. A complementary analysis of PRISM identified 17,946 compound-aneuploidy associations and 16,189 mechanism of action (MOA)-aneuploidy associations. Pathway-altering compounds that selectively reduce viability in cells with aneuploidy profiles were discovered, including an unexpectedly prominent number of glucocorticoid receptor agonists.
Conclusions: This integrated dataset provides a resource for designing therapeutic decision hypotheses, identifying drug-repurposing opportunities, and informing future studies aimed at targeting aneuploidy-induced vulnerabilities in cancer.
Keywords: aneuploidy; cancer; chemotherapy; oncology; precision oncology.
Conflict of interest statement
The authors declare that they have no competing interests. The funders had no role in the design of this study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

