Cross-sectional Study: Diagnostic Accuracy of Next-generation Sequencing in a Tertiary Care Intensive Care Unit
- PMID: 40567315
- PMCID: PMC12186076
- DOI: 10.5005/jp-journals-10071-24987
Cross-sectional Study: Diagnostic Accuracy of Next-generation Sequencing in a Tertiary Care Intensive Care Unit
Abstract
Background and aims: Infectious diseases are a major cause of intensive care unit (ICU) mortality, where rapid pathogen identification is crucial. Traditional culture methods are slow and may miss fastidious organisms. Next-generation sequencing (NGS) offers rapid, comprehensive pathogen detection. This study assessed NGS accuracy compared to culture in a tertiary care ICU in India.
Patients and methods: A retrospective observational analysis of 187 ICU patients with suspected infections was conducted with IRB approval. Paired samples from blood, urine, bronchoalveolar lavage fluid (BALF), cerebrospinal fluid (CSF), and other body fluids underwent NGS and culture testing. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated using culture as the reference. Concordance was also assessed.
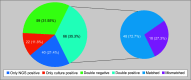
Results: Next-generation sequencing demonstrated a sensitivity of 75%, specificity of 59.6%, PPV of 62.23%, and NPV of 72.84%. It detected pathogens in 56.68% of cases vs 47.06% by culture, identifying 17 atypical organisms in culture-negative cases. Sensitivity was highest in CSF (100%) and BALF (87.5%), while specificity was highest in pleural fluid (100%) and blood (87.5%). Overall concordance was 57.2%.
Conclusion: Next-generation sequencing has improved pathogen detection, identifying organisms missed by culture. High sensitivity across sample types suggests its value in ICU diagnostics. However, lower specificity, high cost, and standardization challenges limit standalone use.
Clinical significance: Next-generation sequencing facilitates an earlier ICU infection diagnosis, allowing for prompt targeted treatment and potentially reducing antimicrobial resistance. However, false positives and cost remain barriers. Combining NGS with conventional culture techniques could improve diagnostic accuracy and patient outcomes in the right subset of patients.
How to cite this article: Sawale M, Raj R, Bhide M, Chanchalani G. Cross-sectional Study: Diagnostic Accuracy of Next-generation Sequencing in a Tertiary Care Intensive Care Unit. Indian J Crit Care Med 2025;29(6):498-503.
Keywords: Culture; Infectious diseases; Intensive care unit; Next-generation sequencing; Pathogen detection.
Copyright © 2025; The Author(s).
Conflict of interest statement
Source of support: Nil Conflict of interest: NoneConflict of interest: None
Figures

References
-
- Gosiewski T, Ludwig-Galezowska AH, Huminska K, Sroka-Oleksiak A, Radkowski P, Salamon D, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method – the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36(2):329–336. doi: 10.1007/s10096-016-2805-7. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
