Next-generation phenotyping of inherited retinal diseases from multimodal imaging with Eye2Gene
- PMID: 40567353
- PMCID: PMC12185311
- DOI: 10.1038/s42256-025-01040-8
Next-generation phenotyping of inherited retinal diseases from multimodal imaging with Eye2Gene
Abstract
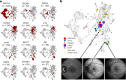
Rare eye diseases such as inherited retinal diseases (IRDs) are challenging to diagnose genetically. IRDs are typically monogenic disorders and represent a leading cause of blindness in children and working-age adults worldwide. A growing number are now being targeted in clinical trials, with approved treatments increasingly available. However, access requires a genetic diagnosis to be established sufficiently early. Critically, the timely identification of a genetic cause remains challenging. We demonstrate that a deep learning algorithm, Eye2Gene, trained on a large multimodal imaging dataset of individuals with IRDs (n = 2,451) and externally validated on data provided by five different clinical centres, provides better-than-expert-level top-five accuracy of 83.9% for supporting genetic diagnosis for the 63 most common genetic causes. We demonstrate that Eye2Gene's next-generation phenotyping can increase diagnostic yield by improving screening for IRDs, phenotype-driven variant prioritization and automatic similarity matching in phenotypic space to identify new genes. Eye2Gene is accessible online (app.eye2gene.com) for research purposes.
Keywords: Genetic testing; Image processing; Molecular medicine; Retinal diseases.
© The Author(s) 2025.
Conflict of interest statement
Competing interestsN.P., W.W., M.M. and A.R.W. are patent holders of PCT/EP2023/076614 filed by UCL Business. I.M. is a cofounder, share-holder and director of Phenopolis Ltd, the software company that developed www.eye2gene.com. N.P. is a cofounder and former share-holder and director of Phenopolis Ltd. A.Y.L. reports grants from Santen, personal fees from Genentech, personal fees from US FDA, personal fees from Johnson and Johnson, grants from Carl Zeiss Meditec, personal fees from Gyroscope, non-financial support from Microsoft and grants from Regeneron, outside the submitted work. M.M. has received consultancy or advisory board fees from MeiraGTx, Janssen Pharmaceuticals, Saliogen and Octant; travel grants from MeiraGTx and Janssen Pharmaceuticals and stock options from MeiraGTx. The other authors declare no competing interests.
Figures



References
-
- Georgiou, M. et al. Phenotyping and genotyping inherited retinal diseases: molecular genetics, clinical and imaging features, and therapeutics of macular dystrophies, cone and cone-rod dystrophies, rod-cone dystrophies, Leber congenital amaurosis, and cone dysfunction syndromes. Prog. Retin. Eye Res.100, 101244 (2024). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
