Automated landmark-based symmetric and standard alignment of skull base structures on CT
- PMID: 40567446
- PMCID: PMC12172739
- DOI: 10.1016/j.ynirp.2025.100260
Automated landmark-based symmetric and standard alignment of skull base structures on CT
Abstract
Introduction: Symmetry and standard alignment are crucial in both clinical interpretation and research on head CT studies. Registration to a standard template is the traditional method for alignment, yet registration does not guarantee precise alignment of any given structure. This study introduces a method for aligning skull base structures while still achieving a standard anterior commissure-posterior commissure (AC-PC)-like orientation on head CT studies using landmarks, specifically the cochleas and nasal bridge.
Methods: A retrospective study was conducted using head CTs from various General Electric scanners. Landmarks were manually annotated, and a 3D U-Net was trained for landmark identification. Landmark-based alignment was then performed on a test dataset and assessed in two different ways: whole head and skull base alignment. Whole head alignment was assessed quantitatively by expert review. Skull base alignment was then assessed at the cochleas, comparing their alignment between this landmark-based technique and registration to a template.
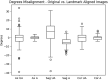
Results: This landmark-based technique significantly improved whole head and skull base alignment of head CT studies. Whole head alignment reduced average deviations of 5, 11, and 4° in the axial, sagittal, and coronal planes to 1, 5, and 2° respectively. Meanwhile, skull base alignment assessed via the cochlea was also improved relative to traditional registration. For the landmark technique, the cochleas were deviated from perfect by a mean of 0.552 and 0.511 mm along the y and z axes compared to 2.110 and 2.506 mm with registration.
Conclusion: This study demonstrates a simple landmark-based technique for aligning the cochleas on head CT studies while approximating whole head AC-PC orientation, which has applications in both clinical and research settings, particularly for studies focused on the skull base.
Keywords: AC-PC line; Alignment; Head CT; Landmark; Registration; U-Net.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
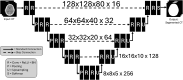



References
-
- Bhattarai R., Panthi S., Yadav G.K., Bhandari S., Acharya R., Sharma A., Shah P.K., Koirala S., Bhattarai M., Gupta M.K., Khanal B. Morphometric analysis of foramen ovale, foramen spinosum, and foramen rotundum of human skull using computed tomography scan: a cross-sectional study. Ann. Med. Surg. 2023;85:1731–1736. doi: 10.1097/MS9.0000000000000609. - DOI - PMC - PubMed
-
- Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. 3D U-net: learning dense volumetric segmentation from sparse annotation. arXiv. 2016 doi: 10.48550/arXiv.1606.06650. - DOI
-
- Consortium M. MONAI: medical open network for AI. 2024. - DOI
-
- Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.-C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S., Miller J.V., Pieper S., Kikinis R. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
