Machine learning driven multi-omics analysis of the genetic mechanisms behind the double-coat fleece formation in Hetian sheep
- PMID: 40567898
- PMCID: PMC12187771
- DOI: 10.3389/fgene.2025.1582244
Machine learning driven multi-omics analysis of the genetic mechanisms behind the double-coat fleece formation in Hetian sheep
Abstract
Introduction: The double-coated fleece is crucial for the adaptability and economic value of Hetian sheep, yet its underlying molecular mechanisms remain largely unexplored.
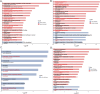
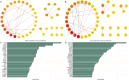
Methods: We integrated genome and transcriptome data from double-coated Hetian sheep and single-coated Chinese Merino sheep. Candidate genes associated with coat fleece type and environmental adaptation were identified using combined selective sweep and differential expression analyses. Subsequent analyses included Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, protein-protein interaction (PPI) network construction, and machine learning-based screening.
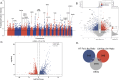
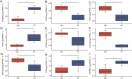
Results: Selective sweep and differential expression analyses identified 101 and 106 candidate genes in Hetian sheep and Chinese Merino sheep, respectively. Enrichment analyses revealed these genes were primarily involved in pathways related to wool growth and energy metabolism. PPI network analysis and machine learning identified IRF2BP2 and EGFR as key functional genes associated with coat fleece type.
Discussion: This study enhances understanding of the genetic mechanisms governing double-coated fleece formation in Hetian sheep. The identification of key genes (IRF2BP2, EGFR) and the methodological approach provide valuable insights for developing machine learning-driven multi-omics selection models in sheep breeding.
Keywords: Chinese merino sheep; Hetian sheep; coat fleece type; machine learning; multi-omics.
Copyright © 2025 Zhang, Li, Xu, Xie, Tang, Zheng, Song, Yu and Di.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Identification of Therapeutic Targets in Autism Spectrum Disorder through CHD8-Notch Pathway Interaction Analysis.PLoS One. 2025 Jun 17;20(6):e0325893. doi: 10.1371/journal.pone.0325893. eCollection 2025. PLoS One. 2025. PMID: 40526590 Free PMC article.
-
Understanding mechanisms of Polygonatum sibiricum-derived exosome-like nanoparticles against breast cancer through an integrated metabolomics and network pharmacology analysis.Front Chem. 2025 Jun 6;13:1559758. doi: 10.3389/fchem.2025.1559758. eCollection 2025. Front Chem. 2025. PMID: 40547857 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

