This is a preprint.
Direct RNA nanopore sequencing reveals rapid RNA modification changes following glucose stimulation of human pancreatic beta-cell lines
- PMID: 40568065
- PMCID: PMC12190750
- DOI: 10.1101/2025.06.12.659352
Direct RNA nanopore sequencing reveals rapid RNA modification changes following glucose stimulation of human pancreatic beta-cell lines
Abstract
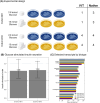
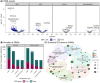
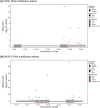
RNA modifications are critical regulators of gene expression and cellular processes; however, the epitranscriptome is less well studied than the epigenome. Here, we studied transcriptome-wide changes in RNA modifications and expression levels in two human pancreatic beta-cell lines, EndoC-BH1 and EndoC-BH3, after one hour of glucose stimulation. Using direct RNA nanopore sequencing (dRNA-seq), we measured N6-methyladenosine (m6A), 5-methylcytosine (m5C), inosine, and pseudouridine concurrently across the transcriptome. We developed a differential RNA modification method and identified 1,697 differentially modified sites (DMSs) across all modifications. These DMSs were largely independent of changes in gene expression levels and enriched in transcripts for type 2 diabetes (T2D) genes. Our study demonstrates how dRNA-seq can be used to detect and quantify RNA modification changes in response to cellular stimuli at the single-nucleotide level and provides new insights into RNA-mediated mechanisms that may contribute to normal beta-cell response and potential dysfunction in T2D.
Conflict of interest statement
LM has received reimbursement of travel or accommodation expenses to speak at Oxford Nanopore Technologies (ONT) conferences. EB is a paid consultant and shareholder of ONT.
Figures

References
-
- Zhang Z. et al. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat. Methods 18, 1213–1222 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
