Systematic evaluation of protein extraction for metaproteomic analysis of marine sediment with high clay content
- PMID: 40568302
- PMCID: PMC12192440
- DOI: 10.1093/ismeco/ycaf074
Systematic evaluation of protein extraction for metaproteomic analysis of marine sediment with high clay content
Abstract
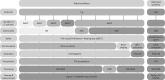
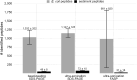
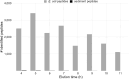
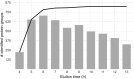
Marine sediments harbor extremely diverse microbial communities that contribute to global biodiversity and play an essential role in the functioning of ecosystems. However, the metaproteome of marine sediments is still poorly understood. The extraction of proteins from environmental samples is still a challenge, especially from marine sediments, due to the complexity of the matrix. Therefore, methods for protein extraction from marine sediments need to be improved. To develop an effective workflow for protein extraction for clayey sediments, we compared, combined and enhanced different protein extraction methods. The workflow presented here includes blocking of protein binding sites on sediment particles with high concentrations of amino acids, effective cell lysis by ultrasonic capture, electro-elution, and simultaneous fractionation of proteins. To test the protocol's efficacy, we added Escherichia coli cells to sediment samples before protein extraction. By using our refined workflow, we were able to identify a comparable number of E. coli proteins from the supplemented sediment to those from pure E. coli cultures. This new protocol will enable future studies to identify active players in clay-rich marine sediments and accurately determine functional biodiversity based on their respective protein complements.
Keywords: Metaproteomics; marine sediment; protein extraction.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures


References
-
- Wang D-Z, Xie Z-X, Zhang S-F. Marine metaproteomics: current status and future directions. J Proteome 2014;97:27–35. - PubMed
LinkOut - more resources
Full Text Sources

