Morphological and molecular identification of the cat flea Ctenocephalides felis from Bangladesh
- PMID: 40568498
- PMCID: PMC12186780
- DOI: 10.5455/javar.2025.l894
Morphological and molecular identification of the cat flea Ctenocephalides felis from Bangladesh
Abstract
Objective: The present study was designed to conduct molecular and morphological identification of cat fleas (Ctenocephalides felis) from Bangladesh along with nucleotide polymorphism and phylogenetic analysis.
Materials and methods: Samples were collected from two hosts (cat and human). The species was identified through morphological studies first, and then DNA was extracted for subsequent molecular analysis. A part of the mitochondrial 16S rRNA gene was amplified by polymerase chain reaction using extracted DNA as a template. The amplified region was sequenced using the Sanger dideoxy method. The sequence was subjected to NCBI BLASTn search. BioEdit and MEGA 11 software were used for multiple sequence alignment (MSA) and generating a phylogenetic tree.
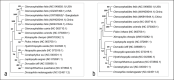
Results: Morphological features such as shape, size, and appendages showed similarity with C. felis. The metatibial formula of chaetotaxy (2-2-2-2-1-3) was confirmed for species-level identification. NCBI BLASTn search showed the highest sequence identity with the available sequence of C. felis such as 99.78% (NC_049858.1) and 99.12% (MW420044.1 and MK941844.1) with 100% query coverage. MSA of C. felis sequences from different geographical distributions show their sequence affinities with each other, and the phylogenetic tree presents their relationship with each other.
Conclusion: Both morphological and molecular studies clearly indicate the identity and confirmation of cat flea (C. felis) from Bangladesh.
Keywords: 16S rRNA; Cat flea; Ctenocephalides felis; ectoparasite; molecular identification; morphological identification.
© The authors.
Conflict of interest statement
The authors declared no potential conflicts of interest.
Figures


References
-
- Moore CO, André MR, Šlapeta J, Breitschwerdt EB. Vector biology of the cat flea Ctenocephalides felis. Trends Parasitol. 2024;40(4):324–37. https://doi.org/10.1016/j.pt.2024.02.006. - PMC - PubMed
-
- Calvani NED, Bell L, Carney A, De La Fuente C, Stragliotto T, Tunstall M, et al. The molecular identity of fleas (Siphonaptera) carrying Rickettsia felis, Bartonella clarridgeiae and Bartonella rochalimae from dogs and cats in Northern Laos. Heliyon. 2020;6(7):4–7. https://doi.org/10.1016/j.heliyon.2020.e04385. - PMC - PubMed
-
- Beliavskaia A, Tan KK, Sinha A, Husin NA, Lim FS, Loong SK, et al. Metagenomics of culture isolates and insect tissue illuminate the evolution of Wolbachia, Rickettsia and Bartonella symbionts in Ctenocephalides spp. fleas. Microb Genom. 2023;9(7):001045. https://doi.org/10.1099/mgen.0.001045. - PMC - PubMed
-
- Hamzaoui B El, Zurita A, Cutillas C, Parola P. Fleas and flea-borne diseases of North Africa. Acta Trop. 2020;211:105627. https://doi.org/10.1016/j.actatropica.2020.105627. - PubMed
-
- Ehlers J, Krüger A, Rakotondranary SJ, Ratovonamana RY, Poppert S, Ganzhorn JU, et al. Molecular detection of Rickettsia spp., Borrelia spp., Bartonella spp. and Yersinia pestis in ectoparasites of endemic and domestic animals in southwest Madagascar. Acta Trop. 2020;205:105339. https://doi.org/10.1016/j.actatropica.2020.105339. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
