Two-dimensional QSAR-driven virtual screening for potential therapeutics against Trypanosoma cruzi
- PMID: 40568639
- PMCID: PMC12188446
- DOI: 10.3389/fchem.2025.1600945
Two-dimensional QSAR-driven virtual screening for potential therapeutics against Trypanosoma cruzi
Abstract
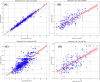
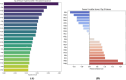
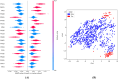
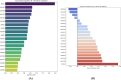
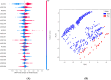
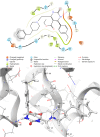
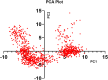
Trypanosoma cruzi is the cause of Chagas disease (CD), a major health issue that affects 6-7 million individuals globally. Once considered a local problem, migration and non-vector transmission have caused it to spread. Efforts to eliminate CD remain challenging due to insufficient awareness, inadequate diagnostic tools, and limited access to healthcare, despite its classification as a neglected tropical disease (NTD) by the WHO. One of the foremost concerns remains the development of safer and more effective anti-Chagas therapies. In our study, we developed a standardized and robust machine learning-driven QSAR (ML-QSAR) model using a dataset of 1,183 Trypanosoma cruzi inhibitors curated from the ChEMBL database to speed up the drug discovery process. Following the calculation of molecular descriptors and feature selection approaches, Support Vector Machine (SVM), Artificial Neural Network (ANN), and Random Forest (RF) models were developed and optimized to elucidate and predict the inhibition mechanism of novel inhibitors. The ANN-driven QSAR model utilizing CDK fingerprints exhibited the highest performance, proven by a Pearson correlation coefficient of 0.9874 for the training set and 0.6872 for the test set, demonstrating exceptional prediction accuracy. Twelve possible inhibitors with pIC50 ≥ 5 were further identified through screening of large chemical libraries using the ANN-QSAR model and ADMET-based filtering approaches. Molecular docking studies revealed that F6609-0134 was the best hit molecule. Finally, the stability and high binding affinity of F6609-0134 were further validated by molecular dynamics simulations and free energy analysis, bolstering its continued assessment as a possible treatment option for Chagas disease.
Keywords: Chagas disease; Trypanosoma cruzi; artificial neural network; machine learning; molecular docking; molecular dynamics; quantitative structure activity relationships; virtual screening.
Copyright © 2025 Maliyakkal, Kumar, Bhowmik, Vishwakarma, Yadav and Mathew.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Integrating traditional QSAR and read-across-based regression models for predicting potential anti-leishmanial azole compounds.Mol Divers. 2025 Aug;29(4):3207-3231. doi: 10.1007/s11030-024-11070-w. Epub 2024 Dec 10. Mol Divers. 2025. PMID: 39653961
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Exploring Type II Diabetes Inhibitors from Genus Daphne Plant-species: An Integrated Computational Study.Comb Chem High Throughput Screen. 2025;28(8):1413-1442. doi: 10.2174/0113862073262227231005074024. Comb Chem High Throughput Screen. 2025. PMID: 38584562
-
Exploring the mechanisms of action of the antimicrobial peptide CZS-5 against Trypanosoma cruzi epimastigotes: insights from metabolomics and molecular dynamics.Parasit Vectors. 2025 Jun 5;18(1):208. doi: 10.1186/s13071-025-06861-5. Parasit Vectors. 2025. PMID: 40474242 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Abras A., Ballart C., Fernández-Arévalo A., Pinazo M.-J., Gascón J., Muñoz C., et al. (2022). Worldwide control and management of Chagas disease in a new era of globalization: a close look at congenital Trypanosoma cruzi infection. Clin. Microbiol. Rev. 35 (2), e0015221. 10.1128/cmr.00152-21 - DOI - PMC - PubMed
-
- Baldi A. (2010). Computational approaches for drug design and discovery: an overview. Syst. Rev. Pharm. 1 (1), 99. 10.4103/0975-8453.59519 - DOI
-
- Breiman L. (2001). Random forests. Mach. Learn. 45 (1), 5–32. 10.1023/A:1010933404324 - DOI
-
- Cao D., Deng Z., Zhu M., Yao Z., Dong J., Zhao R. (2017). Ensemble partial least squares regression for descriptor selection, outlier detection, applicability domain assessment, and ensemble modeling in QSAR/QSPR modeling. J. Chemom. 31 (11). 10.1002/cem.2922 - DOI
LinkOut - more resources
Full Text Sources

