This is a preprint.
Analysis of BRCA1, BRCA2 and PALB2 related Fanconi anemia identifies scope to expand disease phenotypic features and predict breast cancer risk in heterozygotes
- PMID: 40568666
- PMCID: PMC12191086
- DOI: 10.1101/2025.05.25.25327887
Analysis of BRCA1, BRCA2 and PALB2 related Fanconi anemia identifies scope to expand disease phenotypic features and predict breast cancer risk in heterozygotes
Abstract
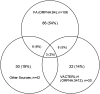
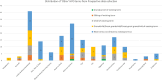
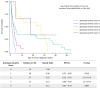
Recessive Fanconi anemia (FA) phenotype is used in classification of BRCA1/FANCS, BRCA2/FANCD1 and PALB2/FANCN variants with respect to dominant hereditary breast-ovarian cancer syndrome. We assessed its utility by examining the spectrum of phenotypes observed in individuals biallelic for BRCA1, BRCA2 or PALB2 pathogenic variants, and exploring the relationship between cancer presentation and allele severity score based on variant molecular features. A data collection instrument comprising 158 Human Phenotype Ontology (HPO) terms was used to document clinical features for individuals with FA from published and/or prospectively collected (total n=172, 43 previously unpublished) phenotypic data. Unique FA-related variants (15 BRCA1, 123 BRCA2, 22 PALB2) were annotated for predicted molecular impact, location, observed splicing or functional impact, and potential in-frame splice rescue. Annotations were used to assign different permutations of allele severity scores, which were assessed for correlation with FA presentation features. The association of BRCA1 and BRCA2 allele severity score with magnitude of breast cancer risk in heterozygotes was evaluated using case-control analysis. Patient-detected features extended beyond the FA ORPHA:84 HPO list, including 84 terms related by hierarchy, and 94 novel terms. Genotype severity score was significantly associated with age at cancer diagnosis in BRCA2 FA individuals (p=1.8×10-8). A similar permutation approach revealed significant differences in magnitude of breast cancer risk according to BRCA1 and BRCA2 allele severity score in heterozygotes. Findings indicate potential to redefine the existing list of FA-related HPO terms, and to use an allele severity scoring approach to predict cancer risk in both FA patients and heterozygotes.
Conflict of interest statement
J.S and J.N. have received research funding from pharma and biotech companies unrelated to this research. T.v.O.H. has received lecture honoraria from AstraZeneca. K.T. has received reimbursement from Merck Sharp and Dolme for work related to Von-Hippel lindau disease. All other authors declare no conflicts of interest.
Figures


References
-
- Mehta P.A., and Ebens C. (2002). Fanconi Anemia. In GeneReviews((R)), Adam M.P., Feldman J., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., and Amemiya A., eds. (University of Washington, Seattle; 1993–2024). - PubMed
-
- Nakano Y., Kuiper R.P., Nichols K.E., Porter C.C., Lesmana H., Meade J., Kratz C.P., Godley L.A., Maese L.D., Achatz M.I., et al. (2024). Update on Recommendations for Cancer Screening and Surveillance in Children with Genomic Instability Disorders. Clin Cancer Res 30, 5009–5020. 10.1158/1078-0432.CCR-24-1098. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
