This is a preprint.
The Landscape of Shared and Divergent Genetic Influences across 14 Psychiatric Disorders
- PMID: 40568675
- PMCID: PMC12191084
- DOI: 10.1101/2025.01.14.25320574
The Landscape of Shared and Divergent Genetic Influences across 14 Psychiatric Disorders
Abstract
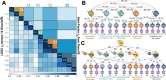
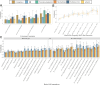
Psychiatric disorders display high levels of comorbidity and genetic overlap1,2. Genomic methods have shown that even for schizophrenia and bipolar disorder, two disorders long-thought to be etiologically distinct3, the majority of genetic signal is shared4. Furthermore, recent cross-disorder analyses have uncovered over a hundred pleiotropic loci shared across eight disorders5. However, the full scope of shared and disorder-specific genetic basis of psychopathology remains largely uncharted. Here, we address this gap by triangulating across a suite of cutting-edge statistical genetic and functional genomic analyses applied to 14 childhood- and adult-onset psychiatric disorders (1,056,201 cases). Our analyses identify and characterize five underlying genomic factors6 that explain the majority of the genetic variance of the individual disorders (~66% on average) and are associated with 268 pleiotropic loci. We observed particularly high levels of polygenic overlap7 and local genetic correlation8 and very few disorder-specific loci9 for two factors defined by: (i) schizophrenia and bipolar disorder ("SB factor"), and by (ii) major depression, PTSD, and anxiety ("internalizing factor"). At the functional level, we applied multiple methods10-12 which demonstrated that the shared genetic signal across the SB factor was substantially enriched in genes expressed in excitatory neurons, whereas the internalizing factor was associated with oligodendrocyte biology. By comparison, the genetic signal shared across all 14 disorders was enriched for broad biological processes (e.g., transcriptional regulation). These results indicate increasing differentiation of biological function at different levels of shared cross-disorder risk, from quite general vulnerability to more specific pathways associated with subsets of disorders. These observations may inform a more neurobiologically valid psychiatric nosology and implicate novel targets for therapeutic developments designed to treat commonly occurring comorbid presentations.
Conflict of interest statement
J.W.S. is a member of the Scientific Advisory Board of Sensorium Therapeutics (with equity) and has received an honorarium for an internal seminar Tempus Labs. K.P.J. is a consultant for Allia Health. A.D.B. has received a speaker fee from Lundbeck. In the past year, S.V.F. received income, potential income, travel expenses continuing education support and/or research support from Aardvark, Aardwolf, AIMH, Akili, Atentiv, Axsome, Genomind, Ironshore, Johnson & Johnson/Kenvue, Kanjo, KemPharm/Corium, Noven, Otsuka, Sky Therapeutics, Sandoz, Supernus, Tris, and Vallon. With his institution, S.V.F. has US patent US20130217707 A1 for the use of sodium-hydrogen exchange inhibitors in the treatment of ADHD. S.V.F. also receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health, Oxford University Press: Schizophrenia: The Facts and Elsevier: ADHD: Non-Pharmacologic Interventions and is Program Director of www.ADHDEvidence.org and www.ADHDinAdults.com.
Figures




References
Publication types
Grants and funding
- R01 MH124851/MH/NIMH NIH HHS/United States
- R01 MH104964/MH/NIMH NIH HHS/United States
- P2C HD042849/HD/NICHD NIH HHS/United States
- R01 MH124847/MH/NIMH NIH HHS/United States
- R01 MH130899/MH/NIMH NIH HHS/United States
- R01 MH123775/MH/NIMH NIH HHS/United States
- U01 MH109514/MH/NIMH NIH HHS/United States
- DP1 DA054394/DA/NIDA NIH HHS/United States
- R01 MH123922/MH/NIMH NIH HHS/United States
- U01 MH109528/MH/NIMH NIH HHS/United States
- U01 MH096296/MH/NIMH NIH HHS/United States
- U01 MH085518/MH/NIMH NIH HHS/United States
- R01 MH136149/MH/NIMH NIH HHS/United States
- U01 MH085520/MH/NIMH NIH HHS/United States
- U01 MH135970/MH/NIMH NIH HHS/United States
- U01 MH085513/MH/NIMH NIH HHS/United States
- U01 MH109499/MH/NIMH NIH HHS/United States
- K08 MH122673/MH/NIMH NIH HHS/United States
- K08 MH135343/MH/NIMH NIH HHS/United States
- U54 GM115516/GM/NIGMS NIH HHS/United States
- F30 MH135712/MH/NIMH NIH HHS/United States
- R21 MH123908/MH/NIMH NIH HHS/United States
- R01 DA054869/DA/NIDA NIH HHS/United States
- U01 MH094411/MH/NIMH NIH HHS/United States
- R01 MH121521/MH/NIMH NIH HHS/United States
- R01 MH121924/MH/NIMH NIH HHS/United States
- R01 MH124873/MH/NIMH NIH HHS/United States
- R01 AG073593/AG/NIA NIH HHS/United States
- U01 MH109536/MH/NIMH NIH HHS/United States
- U01 MH109501/MH/NIMH NIH HHS/United States
- R01 NS128535/NS/NINDS NIH HHS/United States
- P30 AG066614/AG/NIA NIH HHS/United States
- R01 MH112904/MH/NIMH NIH HHS/United States
- U24 MH068457/MH/NIMH NIH HHS/United States
- R01 MH124875/MH/NIMH NIH HHS/United States
- R01 MH123451/MH/NIMH NIH HHS/United States
- R01 MH124871/MH/NIMH NIH HHS/United States
- R01 MH124839/MH/NIMH NIH HHS/United States
- U01 MH085508/MH/NIMH NIH HHS/United States
- U01 MH109532/MH/NIMH NIH HHS/United States
- R01 MH116037/MH/NIMH NIH HHS/United States
- U01 MH109539/MH/NIMH NIH HHS/United States
- U01 MH094421/MH/NIMH NIH HHS/United States
- R01 MH131685/MH/NIMH NIH HHS/United States
- R01 MH106595/MH/NIMH NIH HHS/United States
- U01 MH094432/MH/NIMH NIH HHS/United States
- U01 AR076092/AR/NIAMS NIH HHS/United States
- R01 GM148494/GM/NIGMS NIH HHS/United States
- R01 MH119243/MH/NIMH NIH HHS/United States
- R01 MH120219/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
