The emerging roles of ubiquitin-like modifications in regulating HIV replication and host defense
- PMID: 40568704
- PMCID: PMC12187746
- DOI: 10.3389/fcimb.2025.1593445
The emerging roles of ubiquitin-like modifications in regulating HIV replication and host defense
Abstract
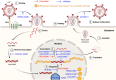
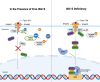
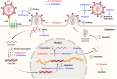
As a post-translational modification (PTM) mechanism analogous to ubiquitination, ubiquitin-like (UBL) modification plays a crucial regulatory role in virus-host interactions. With the increasing discovery of UBL modification types, their roles in diverse biological process, including HIV infection, have gained growing attention. Rather than merely serving as anti-HIV defenses or being exploited by the virus, UBLs often exert dual roles by modulating both host restriction factors and viral proteins, thereby impacting key steps of HIV life cycle, immune evasion, and intracellular signaling. This article summarizes recent advances on the contribution of UBLs in regulating HIV replication and host defense, highlighting their indispensable roles in arms races between HIV and host, aiming to provide a theoretical framework for developing novel therapeutic strategies against HIV-1 targeting virus-host interactions.
Keywords: ATG8ylation; FAT10ylation; HIV-1; ISGylation; NEDDylation; SUMOylation; UBLs.
Copyright © 2025 Yuan, Wang, Sun and Huan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Akbari M., Otterlei M., Peña-Diaz J., Aas P. A., Kavli B., Liabakk N. B., et al. (2004). Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 32, 5486–5498. doi: 10.1093/nar/gkh872 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

