Pyridine-bis(benzimidazole) induces DNA damage at G-quadruplex loci and promotes synthetic lethality with DNA repair inhibition
- PMID: 40568940
- PMCID: PMC12199159
- DOI: 10.1093/nar/gkaf543
Pyridine-bis(benzimidazole) induces DNA damage at G-quadruplex loci and promotes synthetic lethality with DNA repair inhibition
Abstract
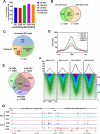
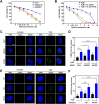
G-quadruplexes (G4s) are noncanonical DNA structures that play key roles in regulating replication, transcription, and genome stability. Here, we investigate the effects of pyridine-bis(benzimidazole) (PyBI), a selective parallel and hybrid G4 stabilizer, on genome stability in cells. Biophysical and biochemical assays confirm PyBI's strong affinity for parallel G4s, leading to replication fork stalling and transcriptional repression of G4-associated oncogenes. Cleavage under targets and tagmentation (CUT&Tag) sequencing reveals a PyBI-induced genome-wide increase in G4 peaks, particularly at promoter and transcription start site regions. This G4 induction and stabilization triggers replication stress, G2/M arrest, and apoptosis. DNA repair pathway profiling shows that PyBI-induced G4 stabilization activates both homologous recombination and nonhomologous end joining (NHEJ), with a predominant role for NHEJ, as indicated by higher 53BP1 colocalization at G4 sites. Genome-wide mapping of PyBI-induced DNA breaks further supports a direct link between G4 stabilization and DNA damage. Moreover, PyBI shows synthetic lethality in DNA repair-deficient contexts, highlighting its therapeutic potential. These findings provide mechanistic insights into the genotoxic effects of PyBI and highlight its potential as a novel anticancer agent targeting G4-mediated genome instability.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures






Similar articles
-
In-depth analysis of the mode of action of resveratrol: genome-wide characterization of G-quadruplex binding properties.Cell Mol Biol Lett. 2025 Jun 20;30(1):74. doi: 10.1186/s11658-025-00747-1. Cell Mol Biol Lett. 2025. PMID: 40542407 Free PMC article.
-
Untargeted CUT&Tag reads are enriched at accessible chromatin and restrict identification of potential G4-forming sequences in G4-targeted CUT&Tag experiments.Nucleic Acids Res. 2025 Jul 19;53(14):gkaf678. doi: 10.1093/nar/gkaf678. Nucleic Acids Res. 2025. PMID: 40682824 Free PMC article.
-
G-quadruplex stabilizer CX-5461 effectively combines with radiotherapy to target α-thalassemia/mental retardation X-linked-deficient malignant glioma.Neuro Oncol. 2025 May 15;27(4):932-947. doi: 10.1093/neuonc/noae248. Neuro Oncol. 2025. PMID: 39570009
-
The G-quadruplex ligand CX-5461: an innovative candidate for disease treatment.J Transl Med. 2025 Apr 18;23(1):457. doi: 10.1186/s12967-025-06473-8. J Transl Med. 2025. PMID: 40251554 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

