Structural and functional studies of the main replication protein NS1 of human parvovirus B19
- PMID: 40568941
- PMCID: PMC12199161
- DOI: 10.1093/nar/gkaf562
Structural and functional studies of the main replication protein NS1 of human parvovirus B19
Abstract
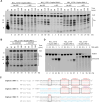
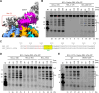
Parvovirus B19 (B19V) is a ubiquitous virus that can infect the majority of human population and cause erythema infectiosum, acute arthropathy, and many other diseases. The main replication protein NS1 plays a critical role in cell cycle arrest, transactivation of viral and host genes, and replication and package of B19V genome. Both DNA nicking and unwinding activities are required for the in vivo function of NS1, but the underlying basis is poorly understood. Here, we report extensive structural and biochemical studies of NS1, showing that NS1 can unwind various types of DNA substrates. The cryo-electron microscopy (cryo-EM) structures reveal the detailed mechanisms for ATP binding and hydrolysis, and DNA binding and unwinding by NS1. In addition to the SF3 HD domain, the C-terminal region is also required for double-stranded DNA (dsDNA) nicking by NS1. Unexpectedly, instead of enhancing, the dsDNA nicking activity of NS1 is negatively regulated by its DNA unwinding ability, suggesting that they likely function in different stages. This study advances our understanding of the structure and function of NS1 and other parvoviral replication proteins, such as the Rep proteins of adeno-associated virus.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

