Testicular somatic and germ cell maturation during rhesus macaque development
- PMID: 40569389
- PMCID: PMC12232671
- DOI: 10.1073/pnas.2419995122
Testicular somatic and germ cell maturation during rhesus macaque development
Abstract
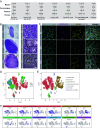
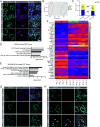
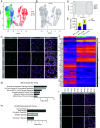
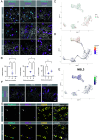
The formation of bilateral testes in animals is critical for puberty, reproductive capacity, and testosterone production across the life course. In humans, testis development begins in embryonic life in the first trimester, with considerable effort focused on the cell and developmental events associated with testis cell specification, leaving limited knowledge on testicular organogenesis during the second and third trimesters. To fill this knowledge gap, we evaluated testicular cell maturation at weeks 5 (W5), W6, W8, W15, and W19 postconception using a rhesus macaque model. Our data identify a major transcriptional change in the somatic cells of the testis (Sertoli cells, interstitial cells and fetal Leydig cells) between W8 and W15, and this is associated with the maturation of seminiferous cords and maturation of PGCs into fetal spermatogonia. Through this work, we identified cellular changes and differential protein expression between W5 and W19 that can be used to holistically define testis development across the time course of embryonic and fetal life. This study provides important insights necessary to recreate the testicular niche from stem cells for biomedical research.
Keywords: gonadal development; rhesus macaque; single-cell RNA-seq; testicular maturation.
Conflict of interest statement
Competing interests statement:ATC is on the Board of Directors for the International Society for Stem Cell Research.
Figures
References
-
- Behringer R. R., Cate R. L., Froelick G. J., Palmiter R. D., Brinster R. L., Abnormal sexual development in transgenic mice chronically expressing müllerian inhibiting substance. Nature 345, 167–170 (1990). - PubMed
MeSH terms
Grants and funding
- P51 OD011092/OD/NIH HHS/United States
- OD011092/HHS | NIH (NIH)
- HD098278/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- UL1TR001881/UC | UCLA | Clinical and Translational Science Institute, University of California, Los Angeles (CTSI)
- UL1 TR001881/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

