Multivariate Screening and Automated Clustering of Macrophage Immunoreactome to Nanoparticles and Photothermal Therapy
- PMID: 40569571
- PMCID: PMC12376647
- DOI: 10.1002/advs.202405860
Multivariate Screening and Automated Clustering of Macrophage Immunoreactome to Nanoparticles and Photothermal Therapy
Abstract
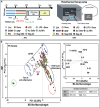
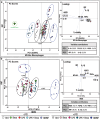
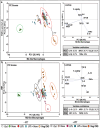
Immunotherapy aims to control the immune system against diseases such as cancer or infections. Nanotechnology is part of the armamentarium to reprogram the immune system in a spatially and temporally controlled manner. However, predicting immune responses using high-throughput tests is challenging due to the immunoreactome's complexity and plasticity. This work presents a fast, machine learning-assisted predictive assay to classify the multifactorial immune responses to any kind of treatments. Engineered human THP-1 monocytes differentiated and polarized into M0, M1, and M2 macrophages are used to monitor nuclear factor Kappa B (NF-kB) and interferon regulatory factor (IRF) pathway activations and gene expression profile in response to metallic nanoparticles (NPs), activated or not by light to induce photothermal therapy (PTT). Principal component analysis (PCA) reveals distinct responses to nanoparticles and the reprogramming toward inflammatory macrophage triggered by PTT. Gold-iron oxide nanoflowers and magnetosomes per se favor polarization toward M2 profile, while light activation shifts this M2-like macrophages toward an M1 phenotype. These findings, confirmed on human blood derived monocytes shed light on the intricate immunomodulatory effects of nanoparticles and PTT on macrophage behavior and provide a basis for an adaptable screening method for the predictive design of therapeutic strategies for immunotherapy in cancer and other diseases.
Keywords: immunotherapy; macrophages; nanoparticles; photothermal therapy.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
