The quest for molecular markers indicating root growth in microbially treated tomato (Solanum lycopersicum) plants
- PMID: 40569659
- PMCID: PMC12199702
- DOI: 10.1093/femsec/fiaf063
The quest for molecular markers indicating root growth in microbially treated tomato (Solanum lycopersicum) plants
Abstract
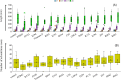
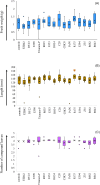
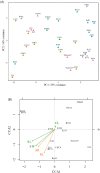
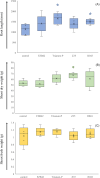
Roots are essential plant organs for anchorage in soil, uptake of water with nutrients, storage of photosynthates, and microbial interactions. More knowledge on microorganisms stimulating root growth is needed to control root development of cultured plants. A marker-assisted approach would facilitate vast screenings of microbes for eventual effects on root development. It was aimed to select for transcripts that report on root growth stimulation at the early tomato plant growth stage upon microbial treatments. Microbially treated tomato (Solanum lycopersicum) plants were cultivated in stone wool slabs and screened for genes that increased or decreased in differential expression upon increased root growth, by RNAseq. Expression of 21 selected genes was measured by quantitative polymerase chain reaction (qPCR) in relation with stimulated root growth, recorded by X-ray microtomography, of microbially treated tomato plants cultivated in stone wool blocks. Two genes were identified of which expression significantly correlated with high measured root length, and for one also with high measured shoot wet and dry weight. The translated products, both involved in modulation of Rubisco activity, were a chloroplast-localized acetyltransferase (SlSNAT2) and a Rubisco activase (Rca). Transcripts whose translated products modulate Rubisco activity can serve as candidates for reporting on early root development upon microbial inoculation.
Keywords: Verrucomicrobium; RNAseq; X-ray microtomography; microbial inoculant; root development; root growth indicator; stone wool; tomato plants.
© The Author(s) 2025. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures
Similar articles
-
Peppermint hydrosol as a novel bio-stimulant promotes growth and antioxidant activity of Solanum lycopersicum L.BMC Plant Biol. 2025 Jul 9;25(1):894. doi: 10.1186/s12870-025-06144-2. BMC Plant Biol. 2025. PMID: 40634846 Free PMC article.
-
Serotonin N-acetyltransferase SlSNAT2 Positively Regulates Tomato Resistance Against Ralstonia solanacearum.Int J Mol Sci. 2025 Jul 7;26(13):6530. doi: 10.3390/ijms26136530. Int J Mol Sci. 2025. PMID: 40650305 Free PMC article.
-
First Report of Meloidogyne enterolobii Infecting Tomato (Solanum lycopersicum) in Texas, United States.Plant Dis. 2025 Jun 26. doi: 10.1094/PDIS-04-25-0740-PDN. Online ahead of print. Plant Dis. 2025. PMID: 40570356
-
Etanercept and infliximab for the treatment of psoriatic arthritis: a systematic review and economic evaluation.Health Technol Assess. 2006 Sep;10(31):iii-iv, xiii-xvi, 1-239. doi: 10.3310/hta10310. Health Technol Assess. 2006. PMID: 16948890
-
Sertindole for schizophrenia.Cochrane Database Syst Rev. 2005 Jul 20;2005(3):CD001715. doi: 10.1002/14651858.CD001715.pub2. Cochrane Database Syst Rev. 2005. PMID: 16034864 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

