Dynamics of Th1/Th17 responses and antimicrobial pathways in leprosy skin lesions
- PMID: 40569678
- PMCID: PMC12404764
- DOI: 10.1172/JCI190736
Dynamics of Th1/Th17 responses and antimicrobial pathways in leprosy skin lesions
Abstract
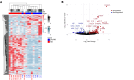
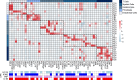
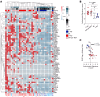
BACKGROUNDReversal reactions (RRs) in leprosy are acute immune episodes marked by inflammation and bacterial clearance, offering a model to study the dynamics of host responses to Mycobacterium leprae. These episodes are often severe and difficult to treat, frequently progressing to permanent disabilities. We aimed to characterize the immune mechanisms and identify antimicrobial effectors during RRs.METHODSWe performed RNA-Seq on paired skin biopsy specimens collected from 9 patients with leprosy before and at RR diagnosis, followed by differential gene expression and functional analysis. A machine-learning classifier was applied to predict membrane-permeabilizing proteins. Antimicrobial activity was assessed in M. leprae-infected macrophages and axenic cultures.RESULTSIn the paired pre-RR and RR biopsy specimens, a 64-gene antimicrobial response signature was upregulated during RR and correlated with reduced M. leprae burden. Predicted upstream regulators included IL-1β, TNF, IFN-γ, and IL-17, indicating activation of both the Th1 and Th17 pathways. A machine-learning classifier identified 28 genes with predicted membrane-permeabilizing antimicrobial activity, including S100A8. Four proteins (S100A7, S100A8, CCL17, and CCL19) demonstrated antimicrobial activity against M. leprae in vitro. Scanning electron microscopy revealed membrane damage in bacteria exposed to these proteins.CONCLUSIONRR is associated with a robust antimicrobial gene program regulated by Th1 and Th17 cytokines. We identified potentially novel host antimicrobial effectors that showed activity against M. leprae, suggesting potential strategies to bolster Th1 and Th17 responses for combating intracellular mycobacterial infections.FUNDINGNIH grants R01 AI022553, R01 AR040312, R01 AR073252, R01 AI166313, R01 AI169526, P50 AR080594, and 4R37 AI052453-21 and National Science Foundation (NSF) grant DMR2325840.
Keywords: Adaptive immunity; Bacterial infections; Immunology; Infectious disease; Innate immunity.
Conflict of interest statement
Figures






References
-
- Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34(3):255–273. - PubMed
-
- Lienhardt C, Fine PE. Type 1 reaction, neuritis and disability in leprosy. What is the current epidemiological situation? Lepr Rev. 1994;65(1):9–33. - PubMed

