Deciphering Probabilistic Species Interaction Networks
- PMID: 40569829
- PMCID: PMC12200232
- DOI: 10.1111/ele.70161
Deciphering Probabilistic Species Interaction Networks
Abstract
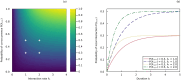
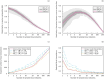
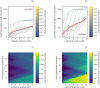
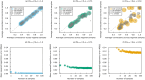
Representing species interactions probabilistically as opposed to deterministically conveys uncertainties in our knowledge of interactions. The sources of uncertainty captured by interaction probabilities depend on the method used to evaluate them: uncertainty of predictive models, subjective assessment of experts, or empirical measurement of interaction spatiotemporal variability. However, guidelines for the estimation and documentation of probabilistic interaction data are lacking. This is concerning because our understanding of interaction probabilities depend on their sometimes elusive definition and uncertainty sources. We review how probabilistic interactions are defined at different spatial scales. These definitions are based on the distinction between the realisation of an interaction at a specific time and space (local networks) and its biological or ecological feasibility (metaweb). Using host-parasite interactions in Europe, we illustrate how these two network representations differ in their statistical properties, specifically: how local networks and metawebs differ in their spatial and temporal scaling of interactions. We present two approaches to inferring binary interactions from probabilistic ones that account for these differences and show that systematic biases arise when directly inferring local networks from metawebs. Our results underscore the importance of more rigorous descriptions of probabilistic species interactions that specify their conditional variables and uncertainty sources.
Keywords: ecological modelling; ecological networks; food webs; host–parasite interactions; metaweb; sampling; spatial scale; species interactions; temporal scale; uncertainty.
Ecology Letters© 2025 The Author(s). Ecology Letters published by John Wiley & Sons Ltd.
Figures
References
-
- Abrego, N. , Roslin T., Huotari T., et al. 2021. “Accounting for Species Interactions Is Necessary for Predicting How Arctic Arthropod Communities Respond to Climate Change.” Ecography 44, no. 6: 885–896.
-
- Angilletta, M. J., Jr. , Steury T. D., and Sears M. W.. 2004. “Temperature, Growth Rate, and Body Size in Ectotherms: Fitting Pieces of a Life‐History Puzzle1.” Integrative and Comparative Biology 44, no. 6: 498–509. - PubMed
-
- Araujo, M. B. , Rozenfeld A., Rahbek C., and Marquet P. A.. 2011. “Using Species Co‐Occurrence Networks to Assess the Impacts of Climate Change.” Ecography 34, no. 6: 897–908.
-
- Bartomeus, I. , Gravel D., Tylianakis J. M., Aizen M. A., Dickie I. A., and Bernard‐Verdier M.. 2016. “A Common Framework for Identifying Linkage Rules Across Different Types of Interactions.” Functional Ecology 30, no. 12: 1894–1903.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

