Natural self-attenuation of pathogenic viruses by deleting the silencing suppressor coding sequence for long-term plant-virus coexistence
- PMID: 40570083
- PMCID: PMC12225820
- DOI: 10.1371/journal.ppat.1013012
Natural self-attenuation of pathogenic viruses by deleting the silencing suppressor coding sequence for long-term plant-virus coexistence
Abstract
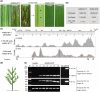
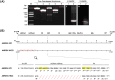
Potyviridae is the largest family of plant-infecting RNA viruses. All members of the family (potyvirids) have single-stranded positive-sense RNA genomes, with polyprotein processing as the expression strategy. The 5'-proximal regions of all potyvirids, except bymoviruses, encode two types of leader proteases: the serine protease P1 and the cysteine protease HCPro. However, their arrangement and sequence composition vary greatly among genera or even species. The leader proteases play multiple important roles in different potyvirid-host combinations, including RNA silencing suppression and virus transmission. Here, we report that viruses in the genus Arepavirus, which encode two HCPro leader proteases in tandem (HCPro1-HCPro2), can naturally lose the coding sequences for these two proteins during infection. Notably, this loss is associated with a shift in foliage symptoms from severe necrosis to mild chlorosis or even asymptomatic infections. Further analysis revealed that the deleted region is flanked by two short repeated sequences in the parental isolates, suggesting that recombination during virus replication likely drives this genomic deletion. Reverse genetic approaches confirmed that the loss of leader proteases weakens RNA silencing suppression and other critical functions. A field survey of areca palm trees displaying varied symptom severity identified a transitional stage in which full-length viruses and deletion mutants coexist in the same tree. Based on these findings, we propose a scenario in which full-length isolates drive robust infections and facilitate plant-to-plant transmission, eventually giving rise to leader protease-less variants that mitigate excessive damage to host trees, allowing long-term coexistence with the perennial host. To our knowledge, this is the first report of potyvirid self-attenuation via coding sequence loss.
Copyright: © 2025 Qin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
A Newly Identified Virus in the Family Potyviridae Encodes Two Leader Cysteine Proteases in Tandem That Evolved Contrasting RNA Silencing Suppression Functions.J Virol. 2020 Dec 9;95(1):e01414-20. doi: 10.1128/JVI.01414-20. Print 2020 Dec 9. J Virol. 2020. PMID: 33055249 Free PMC article.
-
Umbravirus-like RNA viruses are capable of independent systemic plant infection in the absence of encoded movement proteins.PLoS Biol. 2024 Apr 25;22(4):e3002600. doi: 10.1371/journal.pbio.3002600. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38662792 Free PMC article.
-
Enteroviral 3C protease cleaves N4BP1 to impair the host inflammatory response.J Virol. 2025 Jan 31;99(1):e0175824. doi: 10.1128/jvi.01758-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655957 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

