Methods to accelerate PROTAC drug discovery
- PMID: 40570202
- PMCID: PMC12312393
- DOI: 10.1042/BCJ20243018
Methods to accelerate PROTAC drug discovery
Abstract
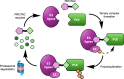
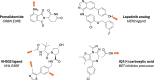
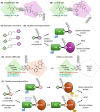
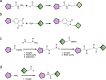
Proteolysis-targeting chimeras (PROTACs) represent a novel and promising modality for probing biological systems, elucidating pharmacological mechanisms, and identifying potential therapeutic leads. The field has made significant strides, as demonstrated by the growing number of PROTACs advancing to clinical trials. Despite this progress, the development of PROTACs faces significant challenges, which is partially due to the heterobivalent nature of this class of molecules. PROTACs must simultaneously bind to a protein of interest and an E3 ubiquitin ligase. This means PROTACs are significantly larger and more complex than conventional small molecules. This complexity impacts their design and synthesis, requiring strategic approaches to create libraries of PROTACs with various combinations of target ligands, linkers, and E3 ligase-recruiting elements. To fully realise the potential of this innovative technology, there is a need for novel approaches to accelerate the development of PROTACs. This review focuses on three critical areas to accelerate PROTAC development: appropriate target selection, modular chemical synthesis, and high-throughput biological evaluation. By appropriate selection of target proteins for degradation, optimizing synthesis methods to handle the complexity of PROTAC molecules, and employing robust high-throughput biological assays to evaluate PROTAC activity, researchers in academia and industry have streamlined the development and increased the success rate of PROTAC-based discovery programmes.
Keywords: DNA-encoded library; click chemistry; direct-to-biology; proteolysis-targeting chimeras; solid-phase synthesis.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

