A lysosomal surveillance response to stress extends healthspan
- PMID: 40571723
- PMCID: PMC12270918
- DOI: 10.1038/s41556-025-01693-y
A lysosomal surveillance response to stress extends healthspan
Abstract
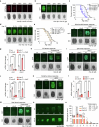
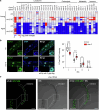
Lysosomes are cytoplasmic organelles central for the degradation of macromolecules to maintain cellular homoeostasis and health. However, how lysosomal activity can be boosted to counteract ageing and ageing-related diseases remains elusive. Here we reveal that silencing specific vacuolar H+-ATPase subunits (for example, vha-6), which are essential for intestinal lumen acidification in Caenorhabditis elegans, extends lifespan by ~60%. This longevity phenotype can be explained by an adaptive transcriptional response typified by induction of a set of transcripts involved in lysosomal function and proteolysis, which we termed the lysosomal surveillance response (LySR). LySR activation is characterized by boosted lysosomal activity and enhanced clearance of protein aggregates in worm models of Alzheimer's disease, Huntington's disease and amyotrophic lateral sclerosis, thereby improving fitness. The GATA transcription factor ELT-2 governs the LySR programme and its associated beneficial effects. Activating the LySR pathway may therefore represent an attractive mechanism to reduce proteotoxicity and, as such, potentially extend healthspan.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures















References
-
- Mutvei, A. P. et al. Balancing lysosome abundance in health and disease. Nat. Cell Biol.25, 1254–1264 (2023). - PubMed
-
- Ballabio, A. & Bonifacino, J. S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol.21, 101–118 (2020). - PubMed
-
- Lawrence, R. E. & Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol.21, 133–142 (2019). - PubMed
-
- Settembre, C. & Perera, R. M. Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology. Nat. Rev. Mol. Cell Biol.25, 223–245 (2024). - PubMed
MeSH terms
Substances
Grants and funding
- ERC-AdG-787702/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- SNSF 31003A_179435; Sinergia CRSII5_202302/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- NRF 2017K1A1A2013124/National Research Foundation of Korea (NRF)
- LT000731/2018-L/Human Frontier Science Program (HFSP)
- PF-19-0232/United Mitochondrial Disease Foundation (UMDF)
LinkOut - more resources
Full Text Sources

