The ACE2 decoy receptor can overcome immune escape by rapid mutating SARS-CoV-2 variants and reduce cytokine induction and clot formation
- PMID: 40571942
- PMCID: PMC12199494
- DOI: 10.1186/s12929-025-01156-4
The ACE2 decoy receptor can overcome immune escape by rapid mutating SARS-CoV-2 variants and reduce cytokine induction and clot formation
Abstract
Background: The COVID-19 pandemic continues to affect the world in 2025. The rapid mutation of SARS-CoV-2 results in breakthrough infections and diminishes the efficacy of vaccines and anti-viral drugs. The severity of the disease varies across different variants, and the underlying mechanisms driving these differences remain unclear. This study explores the relationship between different Spike variants and cytotoxicity, aiming to determine whether the humanized decoy receptor ACE2-Fc can neutralize spikes from diverse variants, offering a solution to overcome rapid mutating SARS-CoV-2 induced immune escape.
Methods: We co-cultured 293 T-ACE2 cells with 293 T cells transfected with various Spike protein variants or used H1650-ACE2 cells transfected with these Spike variants. This allowed us to observe the effects of different Spike mutations, specifically focusing on cell fusion, cytotoxicity, and cytokine release from human peripheral blood mononuclear cells. Flow cytometry is employed to determine if ACE2-Fc can recognize different Spike variants. We also assess the ability of ACE2-Fc to inhibit infection, cell fusion, cytotoxicity, and cytokine release through pseudovirus infections or Spike protein transfections. Additionally, we use actual viruses from SARS-CoV-2 patients to validate the impacts of Spike mutations and the effectiveness of ACE2-Fc. Furthermore, human plasma is utilized to evaluate ACE2-Fc's capability to inhibit Spike-induced clot formation.
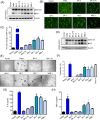
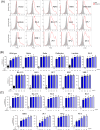
Results: We found that different Spike variants, particularly those with enhancements at the S2' site, increased cell-cell fusion capability, which correlated positively with cytotoxicity and cytokine IL-6 and TNF-α released from PBMCs. ACE2-Fc recognized spikes from wide-range of variants, including wild type, Alpha, Delta, Delta plus, Lambda, BA.2, BA.2.75, BA.5, BF.7, BQ.1, XBB.1, JN.1, KP.2, and KP.3, and effectively prevented these spike-expressing pseudo-viruses from entering host cells. Crucially, ACE2-Fc can prevent spike-induced cell fusion, thereby reducing subsequent cytotoxicity and the release of IL-6 and TNF-α from PBMCs. ACE2-Fc also effectively reduces plasma clot formation induced by trimeric spike proteins.
Conclusions: These findings demonstrated that ACE2-Fc could effectively combat the infection of rapidly mutating SARS-CoV-2, providing a potential solution to overcome immune evasion.
Keywords: ACE2-Fc; Immune escape; SARS-CoV-2; Spike.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Determinants of susceptibility to SARS-CoV-2 infection in murine ACE2.J Virol. 2025 Jun 17;99(6):e0054325. doi: 10.1128/jvi.00543-25. Epub 2025 May 12. J Virol. 2025. PMID: 40353671 Free PMC article.
-
Quantitative characterisation of extracellular vesicles designed to decoy or compete with SARS-CoV-2 reveals differential mode of action across variants of concern and highlights the diversity of Omicron.Cell Commun Signal. 2025 Jul 2;23(1):323. doi: 10.1186/s12964-025-02223-x. Cell Commun Signal. 2025. PMID: 40604989 Free PMC article.
-
Conformational Dynamics and Binding Interactions of SARS-CoV-2 Spike Protein Variants: Omicron, XBB.1.9.2, and EG.5.J Chem Inf Model. 2025 Jul 28;65(14):7651-7667. doi: 10.1021/acs.jcim.5c00308. Epub 2025 Jul 11. J Chem Inf Model. 2025. PMID: 40643983 Free PMC article.
-
The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications.Viruses. 2025 Jul 15;17(7):985. doi: 10.3390/v17070985. Viruses. 2025. PMID: 40733602 Free PMC article. Review.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
References
-
- Liu WJ, Liu P, Lei W, Jia Z, He X, Shi W, Tan Y, Zou S, Wong G, Wang J, et al. Surveillance of SARS-CoV-2 at the huanan seafood market. Nature. 2024;631(8020):402–8. - PubMed
-
- Steiner S, Kratzel A, Barut GT, Lang RM, Aguiar Moreira E, Thomann L, Kelly JN, Thiel V. SARS-CoV-2 biology and host interactions. Nat Rev Microbiol. 2024;22(4):206–25. - PubMed
-
- Lugano D, Kutima B, Kimani M, Sigilai A, Gitonga J, Karani A, Akech D, Karia B, Ziraba AK, Maina A, et al. Evaluation of population immunity against SARS-CoV-2 variants, EG.5.1, FY.4, BA.2.86, JN.1, JN.1.4, and KP.3.1.1 using samples from two health demographic surveillance systems in Kenya. BMC Infect Dis. 2024;24(1):1474. - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

