Vegetation-Driven Changes in Soil Salinity Ions and Microbial Communities Across Tidal Flat Reclamation
- PMID: 40572072
- PMCID: PMC12195035
- DOI: 10.3390/microorganisms13061184
Vegetation-Driven Changes in Soil Salinity Ions and Microbial Communities Across Tidal Flat Reclamation
Abstract
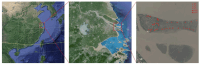
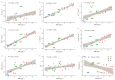
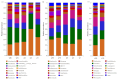
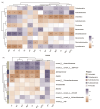
Soil microbes play a vital role in tidal flat ecosystems but are highly susceptible to disturbances from land reclamation. This study investigated the dynamics of bacterial communities and their environmental drivers across a 50-year reclamation chronosequence under three vegetation types (bare flats, reed beds, and rice fields). The results showed that, after 50 years of reclamation, total dissolved salts decreased significantly in vegetated zones, particularly in rice fields, where Cl- dropped by 54.71% and nutrients (SOC, TN, TP) increased substantially. Key ions, including HCO3-, Cl-, and K+, were the primary drivers of microbial community structure, exerting more influence than total salinity (TDS) or pH. Bacterial abundance and diversity increased over time, with rice fields showing the highest values after 50 years. Actinobacteriota and Proteobacteria were positively correlated with HCO3- and K+, while Cl- negatively affected Acidobacteriota. Genus-level analyses revealed that specific taxa, such as Sphingomonas and Gaiella, exhibited ion responses diverging from broader phylum-level patterns, exemplifying niche-specific adaptations to salinity regimes. These findings underscore the pivotal role of vegetation type and individual salinity ions in driving microbial succession during tidal flat reclamation. A phased vegetation strategy, starting with reed colonization and followed by rice cultivation, can enhance soil quality and microbial diversity. This research provides important insights for optimizing vegetation management and ion monitoring in sustainable tidal flat reclamation.
Keywords: reclamation stage; salinity ions; soil bacterial community; tidal flat; vegetation cover type.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
The Impacts of Riparian Vegetation Buffer Strips on Soil Nitrogen Components and Microbial Communities.Environ Manage. 2025 Jul 9. doi: 10.1007/s00267-025-02204-3. Online ahead of print. Environ Manage. 2025. PMID: 40632313
-
The impact of salt-tolerant plants on soil nutrients and microbial communities in soda saline-alkali lands of the Songnen plain.Front Microbiol. 2025 Jun 5;16:1592834. doi: 10.3389/fmicb.2025.1592834. eCollection 2025. Front Microbiol. 2025. PMID: 40539100 Free PMC article.
-
Actinobacteria Emerge as Novel Dominant Soil Bacterial Taxa in Long-Term Post-Fire Recovery of Taiga Forests.Microorganisms. 2025 May 29;13(6):1262. doi: 10.3390/microorganisms13061262. Microorganisms. 2025. PMID: 40572151 Free PMC article.
-
Effects of land use and land cover on soil erosion control in southern China: Implications from a systematic quantitative review.J Environ Manage. 2021 Mar 15;282:111924. doi: 10.1016/j.jenvman.2020.111924. Epub 2021 Jan 9. J Environ Manage. 2021. PMID: 33434792
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
References
-
- Cui L.Q., Liu Y.M., Yan J.L., Hina K., Hussain Q., Qiu T.J., Zhu J.Y. Revitalizing coastal saline-alkali soil with biochar application for improved crop growth. Ecol. Eng. 2022;179:106594. doi: 10.1016/j.ecoleng.2022.106594. - DOI
-
- Cabral R.L., Ferreira T.O., Nóbrega G.N., Barcellos D., Roiloa S.R., Zandavalli R.B., Otero X.L. How do plants and climatic conditions control soil properties in hypersaline tidal flats? Appl. Sci. 2020;10:7624. doi: 10.3390/app10217624. - DOI
-
- Ding B., Bai Y., Guo S., He Z., Wang B., Liu H., Zhai J., Cao H. Effect of irrigation water salinity on soil characteristics and microbial communities in cotton fields in Southern Xinjiang, China. Agronomy. 2023;13:1679. doi: 10.3390/agronomy13071679. - DOI
Grants and funding
- Hu-Nong-Ke-Zhuo 2022[008]/the Outstanding Team Program of Shanghai Academy of Agricultural Science
- Nong-Ke-Jiao-Cha 2024[01]/the Scenario Application-Interdisciplinary Integration and Innovation Pre-research Project of Shanghai Academy of Agricultural Science
- 20025800500/the Domestic Cooperation Program of Shanghai Science and Technology Commission
LinkOut - more resources
Full Text Sources

