Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China
- PMID: 40572085
- PMCID: PMC12195111
- DOI: 10.3390/microorganisms13061197
Population Genomics, Virulence Traits, and Antimicrobial Resistance of Streptococcus suis Isolated in China
Abstract
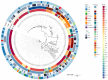
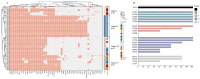
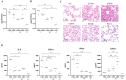
Streptococcus suis is a significant zoonotic pathogen of public health importance. In this study, whole-genome sequencing of 177 isolates of Streptococcus suis, isolated from diseased swine across 15 provinces in China between 2017 and 2019, was performed. A total of 23 serotypes and 28 ST types were identified, with serotypes 2 and 3 comprising 50.8% of the isolates, and sequence types ST353 and ST117 accounting for 23.7%. Clustering analysis based on known virulence-associated factors (VAFs) resulted in the identification of four distinct clusters, and virulence was assessed using animal models, including a unique, highly virulent cluster designated as cluster I. Drug susceptibility testing indicated that 97.7% of the isolates were multidrug-resistant. A total of 26 resistance-associated genes were identified within the genome, 18 of which were associated with integrative and conjugative elements (ICEs) and/or integrative mobilizable elements (IMEs). Nevertheless, our understanding of suis virulence in terms of phylogeny remains incomplete. This study contributes to the understanding of the population structure and genetic characteristics of suis, provides a framework and novel partitioning approach for future investigations into its virulence and pathogenicity, and complements the data on antibiotic resistance.
Keywords: Streptococcus suis; antimicrobial resistance; genomics; virulence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Bojarska A., Molska E., Janas K., Skoczyńska A., Stefaniuk E., Hryniewicz W., Sadowy E. Streptococcus suis in invasive human infections in Poland: Clonality and determinants of virulence and antimicrobial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:917–925. doi: 10.1007/s10096-016-2616-x. - DOI - PMC - PubMed
-
- Dutkiewicz J., Zajac V., Sroka J., Wasinski B., Cisak E., Sawczyn A., Kloc A., Wójcik-Fatla A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II–Pathogenesis. Ann. Agric. Environ. Med. 2018;25:186–203. doi: 10.26444/aaem/85651. - DOI - PubMed
-
- Ferrando M.L., De Greeff A., van Rooijen W.J., Stockhofe-Zurwieden N., Nielsen J., Wichgers Schreur P.J., Pannekoek Y., Heuvelink A., Van Der Ende A., Smith H. Host-pathogen interaction at the intestinal mucosa correlates with zoonotic potential of Streptococcus suis. J. Infect. Dis. 2015;212:95–105. doi: 10.1093/infdis/jiu813. - DOI - PMC - PubMed
-
- Goyette-Desjardins G., Auger J.-P., Xu J., Segura M., Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—An update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014;3:1–20. doi: 10.1038/emi.2014.45. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

