Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments
- PMID: 40572087
- PMCID: PMC12194929
- DOI: 10.3390/microorganisms13061199
Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments
Abstract
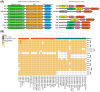
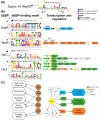
Extreme acidophiles from the Acidithiobacillia class thrive in highly acidic environments where they rely on diverse regulatory mechanisms for adaptation. These mechanisms include sigma factors, transcription factors (TFs), and transcription factor binding sites (TFBS), which control essential pathways. Comparative genomics and bioinformatics analyses identified sigma factors and TFs in Acidithiobacillia, showing similarities but key differences from reference neutrophiles. This study highlights sigma54-dependent one- and two-component systems that are crucial for survival in energy acquisition from sulfur compounds and hydrogen as well as nutrient assimilation. Furthermore, the data suggested evolutionary divergence in regulatory elements distinguishes S-oxidizing from Fe-S-oxidizing members of Acidithiobacillia. Conservation of gene clusters, synteny, and phylogenetic analyses supported the expected phenotypes in each species. Notable examples include HupR's role in hydrogenase-2 oxidation in Fe-S-oxidizers, TspR/TspS regulation of the sulfur oxidation complex, and FleR/FleS control of flagellar motility in S-oxidizers. These regulatory mechanisms act as master controllers of bacterial activity, reflecting adaptation to distinct metabolic needs within Acidithiobacillia.
Keywords: Acidithiobacillia; RpoN; enhancer-binding proteins; hydrogen; motility; nitrogen metabolism; sigma-factors; sulfur metabolism.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Comparative genomics sheds light on transcription factor-mediated regulation in the extreme acidophilic Acidithiobacillia representatives.Res Microbiol. 2024 Jan-Feb;175(1-2):104135. doi: 10.1016/j.resmic.2023.104135. Epub 2023 Sep 9. Res Microbiol. 2024. PMID: 37678513
-
Chemoautotrophy in subzero environments and the potential for cold-adapted Rubisco.Appl Environ Microbiol. 2025 Jun 18;91(6):e0060425. doi: 10.1128/aem.00604-25. Epub 2025 May 30. Appl Environ Microbiol. 2025. PMID: 40444981 Free PMC article.
-
Sulfide Oxidation Products Support Microbial Metabolism at Interface Environments in a Marine-Like Serpentinizing Spring in Northern California.Geobiology. 2025 Jul-Aug;23(4):e70026. doi: 10.1111/gbi.70026. Geobiology. 2025. PMID: 40563104 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
LinkOut - more resources
Full Text Sources
Miscellaneous

