Distribution of Genetic Determinants Associated with CRISPR-Cas Systems and Resistance to Antibiotics in the Genomes of Archaea and Bacteria
- PMID: 40572209
- PMCID: PMC12195208
- DOI: 10.3390/microorganisms13061321
Distribution of Genetic Determinants Associated with CRISPR-Cas Systems and Resistance to Antibiotics in the Genomes of Archaea and Bacteria
Abstract
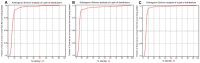
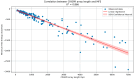
The CRISPR-Cas system represents an adaptive immune mechanism found across diverse Archaea and Bacteria, allowing them to defend against invading genetic elements such as viruses and plasmids. Despite its broad distribution, the prevalence and complexity of CRISPR-Cas systems differ significantly between these domains. This study aimed to characterize and compare the genomic distribution, structural features, and functional implications of CRISPR-Cas systems and associated antibiotic resistance genes in 30 archaeal and 30 bacterial genomes. Through bioinformatic analyses of CRISPR arrays, cas gene architectures, direct repeats (DRs), and thermodynamic properties, we observed that Archaea exhibit a higher number and greater complexity of CRISPR loci, with more diverse cas gene subtypes exclusively of Class 1. Bacteria, in contrast, showed fewer CRISPR loci, comprising a mix of Class 1 and Class 2 systems, with Class 1 representing the majority (~75%) of the detected systems. Notably, Bacteria lacking CRISPR-Cas systems displayed a higher prevalence of antibiotic resistance genes, suggesting a possible inverse correlation between the presence of these immune systems and the acquisition of such genes. Phylogenetic and thermodynamic analyses further highlighted domain-specific adaptations and conservation patterns. These findings support the hypothesis that CRISPR-Cas systems play a dual role: first, as a defense mechanism preventing the integration of foreign genetic material-reflected in the higher complexity and diversity of CRISPR loci in Archaea-and second, as a regulator of horizontal gene transfer, evidenced by the lower frequency of antibiotic resistance genes in organisms with active CRISPR-Cas systems. Together, these results underscore the evolutionary and functional diversification of CRISPR-Cas systems in response to environmental and selective pressures.
Keywords: Archaea and Bacteria; CRISPR-Cas; horizontal gene transfer; resistance to antibiotics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
In silico and in vitro comparative analysis of 79 Acinetobacter baumannii clinical isolates.Microbiol Spectr. 2025 Jul;13(7):e0284924. doi: 10.1128/spectrum.02849-24. Epub 2025 May 16. Microbiol Spectr. 2025. PMID: 40377313 Free PMC article.
-
Horizontal Gene Transfer and CRISPR Targeting Drive Phage-Bacterial Host Interactions and Coevolution in "Pink Berry" Marine Microbial Aggregates.Appl Environ Microbiol. 2023 Jul 26;89(7):e0017723. doi: 10.1128/aem.00177-23. Epub 2023 Jul 5. Appl Environ Microbiol. 2023. PMID: 37404190 Free PMC article.
-
Defense systems and mobile elements in Staphylococcus haemolyticus: a genomic view of resistance dissemination.Microb Pathog. 2025 Sep;206:107808. doi: 10.1016/j.micpath.2025.107808. Epub 2025 Jun 12. Microb Pathog. 2025. PMID: 40516885
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Prophylactic antibiotics for preventing gram-positive infections associated with long-term central venous catheters in adults and children receiving treatment for cancer.Cochrane Database Syst Rev. 2021 Oct 7;10(10):CD003295. doi: 10.1002/14651858.CD003295.pub4. Cochrane Database Syst Rev. 2021. PMID: 34617602 Free PMC article.
References
-
- Barer M., Harwood C. Bacterial Viability and Culturability. Adv. Microb. Physiol. 1999;41:93–137. - PubMed
LinkOut - more resources
Full Text Sources

