OmpR Indirectly Regulates Biosynthesis of Xenocoumacin 1 in Xenorhabdus nematophila
- PMID: 40572248
- PMCID: PMC12196045
- DOI: 10.3390/microorganisms13061360
OmpR Indirectly Regulates Biosynthesis of Xenocoumacin 1 in Xenorhabdus nematophila
Abstract
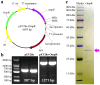
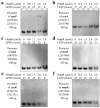
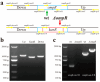
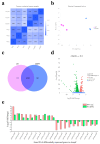
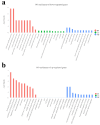
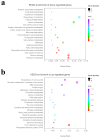
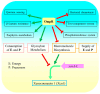
Xenorhabdus nematophila has excellent potential for application in both medicine and agriculture due to its various active secondary metabolites. The transcriptional regulator OmpR negatively regulates Xenocoumacin 1 (Xcn1), which has wide antimicrobial activity. Here, we expressed and purified OmpR and verified its binding activities to promoters via an electrophoretic mobility shift assay. RNA sequencing was used to analyze the relevance and difference of differentially expressed genes between X. nematophila and its mutant ΔompR. Compared with the WT, 1127 differentially expressed genes were found in ΔompR, while 4150 co-expressed genes were detected. RT-qPCR data validated the RNA-seq results with 20 randomly selected genes. OmpR positively regulates the process of porphyrin metabolism, quorum sensing, β-Lactam resistance and glyoxylate and dicarboxylate metabolism, while negatively regulating the phosphotransferase system, two-component system and bacterial chemotaxis. OmpR indirectly regulates the biosynthesis of Xcn1 by positively regulating the process of glyoxylate metabolism, which consumes energy and precursors, and negatively regulates biomacromolecules biosynthesis, which provides energy and precursors. Overall, this work revealed the indirect effects of OmpR on the biosynthesis of Xcn1, serving as a foundation for future research into the intricate regulatory network of X. nematophila.
Keywords: EMSA; OmpR; Xenorhabdus nematophila; transcriptome; xenocoumacin 1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
CpxR negatively regulates the production of xenocoumacin 1, a dihydroisocoumarin derivative produced by Xenorhabdus nematophila.Microbiologyopen. 2019 Feb;8(2):e00674. doi: 10.1002/mbo3.674. Epub 2018 Jun 11. Microbiologyopen. 2019. PMID: 29888873 Free PMC article.
-
Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila.Mol Microbiol. 2009 Sep;73(5):938-49. doi: 10.1111/j.1365-2958.2009.06817.x. Epub 2009 Aug 4. Mol Microbiol. 2009. PMID: 19682255
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Adeolu M., Alnajar S., Naushad S., Gupta R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016;66:5575–5599. - PubMed
-
- Glatter T., Huber M., Lütticke A.L., Papenfort K., Stinear T.P., Bode H.B., Pidot S.J., Tobias N.J., Cai X., Neubacher N. Symbiosis, virulence and natural-product biosynthesis in entomopathogenic bacteria are regulated by a small RNA. Nat. Microbiol. 2020;5:1481–1489. doi: 10.1038/s41564-020-00797-5. - DOI - PMC - PubMed
-
- Hughes D., Forst S., Givaudan A., Gaudriault S., Lanois A., Campagne J.-M., Midrier C., Villain-Guillot P., Sarciaux M., Racine E., et al. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol. Cell. 2018;70:83–94.e7. doi: 10.1016/j.molcel.2018.03.001. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

