Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing
- PMID: 40572261
- PMCID: PMC12196432
- DOI: 10.3390/microorganisms13061373
Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing
Abstract
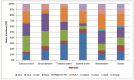
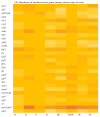
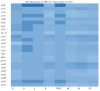
Consumption of contaminated drinking water is known to cause waterborne diseases such as diarrhoea, dysentery, typhoid, and hepatitis. This study applied whole-genome sequencing (WGS) to detect, identify, compare, and profile diarrhoeagenic pathogens (Vibrio cholerae, Shiga toxin-producing Escherichia coli, and Escherichia coli O157:H7) from 3168 water samples and 135 faecal samples (human and animal). Culture-based methods, MALDI-TOF mass spectrometry, and PCR were employed prior to WGS for identification of pathogens. Culture-based results revealed high presumptive prevalence of STEC (40.2%), V. cholerae (37.1%), and E. coli O157:H7 (22.7%). The MALDI-TOF confirmed 555 isolates with V. cholerae identified as Vibrio albensis. Shiga toxin-producing Escherichia coli (STEC) was more prevalent in wastewater (60%), treated water (54.1%), and groundwater (36.8%). PCR detected 46.4% of virulence genes from the water isolates and 66% of virulence genes from the STEC stool isolates. WGS also revealed STEC (92.9%) as the most prevalent species and found common virulence (e.g., hcp1/tssD1 and hlyE) and resistance (e.g., acrA and baeR) genes in all three types of samples. Five resistance and thirteen virulence genes overlapped among treated water and stool isolates. These findings highlight the diarrhoeagenic pathogens' public health risk in water sources and underscore the need for better water quality monitoring and treatment standards.
Keywords: MALDI-TOF MS; conventional PCR; rural communities; water sources; whole-genome sequencing (WGS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- WHO . Guidelines for Drinking Water Quality: Fourth Edition, Incoporating the First and Second Addenda. WHO; Geneva, Switzerland: 2022. - PubMed
-
- Edokpayi J.N., Rogawski E.T., Kahler D.M., Hill C.L., Reynolds C., Nyathi E., Smith J.A., Odiyo J.O., Samie A., Bessong P., et al. Challenges to sustainable safe drinking water: A case study of water quality and use across seasons in rural communities in Limpopo Province, South Africa. Water. 2018;10:159. doi: 10.3390/w10020159. - DOI - PMC - PubMed
-
- Amatobi D.A., Agunwamba J.C. Improved quantitative microbial risk assessment (QMRA) for drinking water sources in developing countries. Appl. Water Sci. 2022;12:49. doi: 10.1007/s13201-022-01569-8. - DOI
-
- Karama M., Mainga A.O., Cenci-Goga B.T., Malahlela M., El-Ashram S., Kalake A. Molecular profiling and antimicrobial resistance of Shiga toxin-producing Escherichia coli O26, O45, O103, O121, O145 and O157 isolates from cattle on cow-calf operations in South Africa. Sci. Rep. 2019;9:11930. doi: 10.1038/s41598-019-47948-1. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

