Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru
- PMID: 40572262
- PMCID: PMC12196064
- DOI: 10.3390/microorganisms13061374
Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru
Abstract
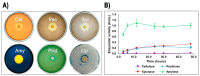
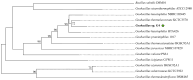
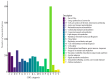
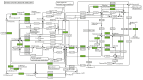
The genome of Geobacillus sp. G4, a thermophilic bacterium isolated from a geothermal field in Peru, was sequenced and analyzed to evaluate its taxonomic and biotechnological potential. This strain exhibits optimal growth at temperatures between 50 and 70 °C and at a pH range of 6.0-7.5. Phenotypic assays demonstrated extracellular enzymatic activities, including amylases, cellulases, pectinases, and xylanases, highlighting its potential for efficient polysaccharide degradation. The assembled genome comprises approximately 3.4 Mb with a G+C content of 52.59%, containing 3,490 genes, including coding sequences, rRNAs, and tRNAs. Functional annotation revealed genes associated with key metabolic pathways such as glycogen and trehalose biosynthesis, indicating adaptation to carbohydrate-rich environments. Phylogenetic analyses based on ANI and dDDH values identified Geobacillus thermoleovorans KCTC3570 as its closest relative, suggesting a strong evolutionary relationship. Additionally, the genome harbors gene clusters for secondary metabolites such as betalactone and fengycin, suggesting potential industrial and pharmaceutical applications, including bioremediation. The identification of antibiotic resistance genes, specifically those conferring glycopeptide resistance, underscores their relevance for antimicrobial resistance studies. The presence of enzymes like amylases and pullulanase further emphasizes its biotechnological potential, particularly in starch hydrolysis and biofuel production. Overall, this research highlights the significant potential of Geobacillus species as valuable sources of thermostable enzymes and biosynthetic pathways for industrial applications.
Keywords: bacterial thermophily; biotechnological potential; industrial enzymes; phylogenomic analysis.
Conflict of interest statement
The authors mutually agree on this submission as it is in the present format; there are no conflicts of interest, and neither human nor animal samples were used during the study. Credit is given to the authors who contributed, and the funding agencies are acknowledged.
Figures





Similar articles
-
Genomic description of Microbacterium mcarthurae sp. nov., a bacterium collected from the International Space Station that exhibits unique antimicrobial-resistant and virulent phenotype.mSystems. 2025 Jun 17;10(6):e0053725. doi: 10.1128/msystems.00537-25. Epub 2025 May 20. mSystems. 2025. PMID: 40391897 Free PMC article.
-
Comprehensive genomic analysis of Bacillus subtilis and Bacillus paralicheniformis associated with the pearl millet panicle reveals their antimicrobial potential against important plant pathogens.BMC Plant Biol. 2024 Mar 18;24(1):197. doi: 10.1186/s12870-024-04881-4. BMC Plant Biol. 2024. PMID: 38500040 Free PMC article.
-
Whole genome sequence of petroleum hydrocarbon degrading novel strain Microbacter sp. EMBS2025 isolated from Chilika Lake, Odisha, India.Sci Rep. 2025 Jul 31;15(1):27961. doi: 10.1038/s41598-025-13545-8. Sci Rep. 2025. PMID: 40745011 Free PMC article.
-
Intravenous magnesium sulphate and sotalol for prevention of atrial fibrillation after coronary artery bypass surgery: a systematic review and economic evaluation.Health Technol Assess. 2008 Jun;12(28):iii-iv, ix-95. doi: 10.3310/hta12280. Health Technol Assess. 2008. PMID: 18547499
-
Dietary interventions for recurrent abdominal pain in childhood.Cochrane Database Syst Rev. 2017 Mar 23;3(3):CD010972. doi: 10.1002/14651858.CD010972.pub2. Cochrane Database Syst Rev. 2017. PMID: 28334433 Free PMC article.
References
-
- Nazina T.N., Tourova T.P., Poltaraus A.B., Novikova E.V., Grigoryan A.A., Ivanova A.E., Lysenko A.M., Petrunyaka V.V., Osipov G.A., Belyaev S.S., et al. Taxonomic Study of Aerobic Thermophilic Bacilli: Descriptions of Geobacillus subterraneus Gen. Nov., Sp. Nov. and Geobacillus uzenensis Sp. Nov. from Petroleum Reservoirs and Transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 2001;51:433–446. doi: 10.1099/00207713-51-2-433. - DOI - PubMed
-
- LPSN—List of Prokaryotic Names with Standing in Nomenclature. [(accessed on 30 March 2025)]. Available online: https://lpsn.dsmz.de/
-
- Coorevits A., Dinsdale A.E., Halket G., Lebbe L., de Vos P., van Landschoot A., Logan N.A. Taxonomic Revision of the Genus Geobacillus: Emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. Thermoglucosidans (Nom. Corrig., Formerly ‘Thermoglucosidasius’); Transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. Int. J. Syst. Evol. Microbiol. 2012;62:1470–1485. doi: 10.1099/ijs.0.030346-0. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

