ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance
- PMID: 40572275
- PMCID: PMC12196164
- DOI: 10.3390/microorganisms13061387
ESBL-Producing Escherichia coli and Klebsiella pneumoniae Exhibit Divergent Paths During In-Human Evolution Towards Carbapenem Resistance
Abstract
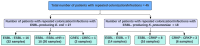
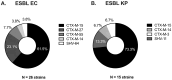
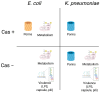
Treatment of infections caused by ESBL-producing Escherichia coli (EC) and Klebsiella pneumoniae (KP) with carbapenem antibiotics can lead to the development of carbapenem resistance over time through the acquisition of porin mutations and plasmids bearing blaKPC. However, the impact of genetic background and the presence of CRISPR-Cas systems on the evolutionary path towards carbapenem resistance in EC and KP has yet to be investigated. The in-human evolution following repeated carbapenem treatment among ESBL-producing Escherichia coli (EC) and Klebsiella pneumoniae (KP) clinical pairs (n = 45 pairs) was examined to determine the relationship between strain genetic background (MLST, CRISPR-Cas) and the evolved genetic mutations related to resistance, virulence, and metabolism by whole genome sequencing. ST131 and ST258 were predominant among seven distinct STs in EC (70%, 19/27) and 11 STs in KP (33%, 6/18), respectively. Complete CRISPR-Cas systems were present in 22% EC (6/27) and 27.8% (5/18) KP pairs, but none in strains belonging to ST131 or ST258; partial loss of CRISPR-Cas was associated with increased carbapenem resistance. Porin, virulence, and metabolism-related genetic mutations were present on the chromosome in both the EC and KP evolved strains, but their presence was differentially associated with the CRISPR-Cas system. Future research on the role of antibiotic exposure in the species-specific resistance evolution of the Enterobacterales could guide antimicrobial stewardship efforts.
Keywords: CRISPR-Cas; ESBL-producing Enterobacterales; carbapenem resistance; genetic evolution.
Conflict of interest statement
The authors declare that this study received funding from Merck & Co. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. The authors declare no conflicts of interest.
Figures


Similar articles
-
Clinical and Antibiotic Resistance Features for Extended-Spectrum Betalactamase-Producing Escherichia coli and Klebsiella pneumoniae Bloodstream Infections and Predictors of Poor Prognosis in Neonatal Patients.Infect Drug Resist. 2025 Aug 5;18:3907-3918. doi: 10.2147/IDR.S530585. eCollection 2025. Infect Drug Resist. 2025. PMID: 40787226 Free PMC article.
-
CRISPR-Cas Dynamics in Carbapenem-Resistant and Carbapenem-Susceptible Klebsiella pneumoniae Clinical Isolates from a Croatian Tertiary Hospital.Pathogens. 2025 Jun 19;14(6):604. doi: 10.3390/pathogens14060604. Pathogens. 2025. PMID: 40559612 Free PMC article.
-
Dissemination of KPC-2-producing carbapenem-resistant Klebsiella pneumoniae ST792 in Southern China.Front Microbiol. 2025 Jun 6;16:1580739. doi: 10.3389/fmicb.2025.1580739. eCollection 2025. Front Microbiol. 2025. PMID: 40547793 Free PMC article.
-
The application of the CRISPR-Cas system in Klebsiella pneumoniae infections.Mol Biol Rep. 2025 Jul 29;52(1):766. doi: 10.1007/s11033-025-10861-0. Mol Biol Rep. 2025. PMID: 40731176 Review.
-
Impact of COVID-19 pandemic on multidrug resistant gram positive and gram negative pathogens: A systematic review.J Infect Public Health. 2023 Mar;16(3):320-331. doi: 10.1016/j.jiph.2022.12.022. Epub 2022 Dec 31. J Infect Public Health. 2023. PMID: 36657243 Free PMC article.
References
-
- Centers for Disease Control and Prevention (USA) Antibiotic Resistance Threats in the United States, 2019. Centers for Disease Control and Prevention (USA); Atlanta, GA, USA: 2019. [(accessed on 4 October 2024)]. Available online: https://stacks.cdc.gov/view/cdc/82532. - DOI
-
- Centers for Disease Control and Prevention (USA) COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. Centers for Disease Control and Prevention (USA); Atlanta, GA, USA: 2022. [(accessed on 4 October 2024)]. Available online: https://stacks.cdc.gov/view/cdc/117915. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

