Metagenomic Investigation of Pathogenic RNA Viruses Causing Diarrhea in Sika Deer Fawns
- PMID: 40573397
- PMCID: PMC12197501
- DOI: 10.3390/v17060803
Metagenomic Investigation of Pathogenic RNA Viruses Causing Diarrhea in Sika Deer Fawns
Abstract
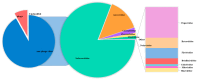
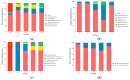
Diarrhea is a common disease in sika deer. The causes of diarrhea in sika deer are complex and involve a variety of pathogens. Additionally, new virulent pathogens are continuously emerging, which poses a serious threat to deer's health and particularly affects fawns' survival rate. In the present study, feces samples were collected from fawns with diarrhea in Jilin Province, in the northeast of China. The viral communities were investigated using the metagenomic method. Viral metagenome data revealed that the viruses in the fecal samples were mainly from 21 families in 14 orders. The major viruses in high abundance were astrovirus, rotavirus, coronavirus, and bovine viral diarrhea virus. In addition, a large number of phages, which mainly belonged to the family Siphoviridae, were identified. Then, the known causative virus species were investigated via RT-qPCR. The results showed that the infection rates of bovine coronavirus, bovine rotavirus, and bovine viral diarrhea virus were 59.44%, 58.89%, and 21.67%, respectively, and mixed infections were commonly seen in the samples. A bovine rotavirus strain was successfully isolated from the positive samples. Whole-genome sequencing revealed that the genotype of the strain was G6-P[1]-I2-R2-C2-M2-A3-N2-T6-E2-H3, indicating the recombination of rotavirus. This study revealed the profiles and characteristics of viruses that cause sika deer diarrhea, which will be helpful for understanding diarrhea diseases in sika deer.
Keywords: rotavirus; sika deer; viral metagenomics; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



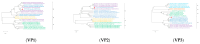
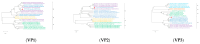

Similar articles
-
Metagenomic insights into the complex viral composition of the enteric RNA virome in healthy and diarrheic calves from Ethiopia.Virol J. 2025 Jun 7;22(1):188. doi: 10.1186/s12985-025-02821-8. Virol J. 2025. PMID: 40483486 Free PMC article.
-
Detection and characterization of bovine coronavirus and rotavirus in calves in Ethiopia.BMC Vet Res. 2025 Feb 28;21(1):122. doi: 10.1186/s12917-025-04563-9. BMC Vet Res. 2025. PMID: 40022093 Free PMC article.
-
Complexity of Diarrhea-Associated Viruses in Stunted Pigs Identified by Viral Metagenomics.Transbound Emerg Dis. 2025 Jun 18;2025:1974716. doi: 10.1155/tbed/1974716. eCollection 2025. Transbound Emerg Dis. 2025. PMID: 40567259 Free PMC article.
-
Prevalence of bovine rotavirus among cattle in mainland China: A meta-analysis.Microb Pathog. 2022 Sep;170:105727. doi: 10.1016/j.micpath.2022.105727. Epub 2022 Aug 19. Microb Pathog. 2022. PMID: 35988882
-
A systematic review on current approaches in bat virus discovered between 2018 and 2022.J Virol Methods. 2024 Sep;329:115005. doi: 10.1016/j.jviromet.2024.115005. Epub 2024 Aug 10. J Virol Methods. 2024. PMID: 39128772
References
-
- Troeger C.E., Khalil I.A., Blacker B.F., Biehl M.H., Albertson S.B., Zimsen S.R.M., Rao P.C., Abate D., Ahmadi A., Ahmed M.L.C., et al. Quantifying risks and interventions that have affected the burden of diarrhoea among children younger than 5 years: An analysis of the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2020;20:37–59. doi: 10.1016/S1473-3099(19)30401-3. - DOI - PMC - PubMed
-
- Pop M., Walker A.W., Paulson J., Lindsay B., Antonio M., Hossain M.A., Oundo J., Tamboura B., Mai V., Astrovskaya I., et al. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76. doi: 10.1186/gb-2014-15-6-r76. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

