Novel Viral Sequences in a Patient with Cryptogenic Liver Cirrhosis Revealed by Serum Virome Sequencing
- PMID: 40573403
- PMCID: PMC12197384
- DOI: 10.3390/v17060812
Novel Viral Sequences in a Patient with Cryptogenic Liver Cirrhosis Revealed by Serum Virome Sequencing
Abstract
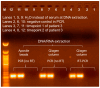
Clinical studies indicate the etiology of liver disease to be unknown in 5% to 30% of patients. A long-standing hypothesis is the existence of unknown viruses beyond hepatitis A through E virus. We conducted serum virome sequencing in nine patients with cryptogenic liver disease and identified eight contigs that could not be annotated. One was determined to be a contaminant, while two of seven contigs from an individual (Patient 3) were validated by reverse transcription and polymerase chain reaction (RT-PCR) and Sanger sequencing. The possibility of contamination was completely excluded through PCR, with templates extracted using different methods from samples taken at different time points. One of the contigs, Seq260, was characterized as negative-sense single-stranded DNA via enzymatic digestion and genome walking. Digital-droplet PCR revealed the copy number of Seq260 to be low: 343 copies/mL. Seq260-based nested PCR screening was negative in 200 blood donors and 225 patients with liver disease with/without known etiologies. None of the seven contigs from Patient 3 was mapped onto 118,713 viral metagenomic data. Conclusively, we discovered seven unknown contigs from a patient with cryptogenic liver cirrhosis. These sequences are likely from a novel human virus with a negative-sense, linear single-stranded DNA genome.
Keywords: cryptogenic cirrhosis; cryptogenic liver disease; etiology; hepatitis virus; virome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

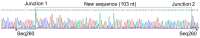


References
-
- Rane S.V., Jain S., Debnath P., Deshmukh R., Nair S., Chandnani S., Kamat R., Rathi P. A comparative study of uncomplicated acute non-A-E hepatitis with acute viral hepatitis and acute onset autoimmune hepatitis. Indian J. Gastroenterol. 2024;43:443–451. doi: 10.1007/s12664-023-01474-1. - DOI - PubMed
-
- Nagaoki Y., Hyogo H., Ando Y., Kosaka Y., Uchikawa S., Nishida Y., Teraoka Y., Morio K., Fujino H., Ono A., et al. Increasing incidence of non-HBV- and non-HCV-related hepatocellular carcinoma: Single-institution 20-year study. BMC Gastroenterol. 2021;21:306. doi: 10.1186/s12876-021-01884-5. - DOI - PMC - PubMed
-
- Ganger D.R., Rule J., Rakela J., Bass N., Reuben A., Stravitz R.T., Sussman N., Larson A.M., James L., Chiu C., et al. Acute liver failure of indeterminate etiology: A comprehensive systematic approach by an expert committee to establish causality. Am. J. Gastroenterol. 2018;113:1319. doi: 10.1038/s41395-018-0160-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

