Synergizing Attribute-Guided Latent Space Exploration (AGLSE) with Classical Molecular Simulations to Design Potent Pep-Magnet Peptide Inhibitors to Abrogate SARS-CoV-2 Host Cell Entry
- PMID: 40573419
- PMCID: PMC12197576
- DOI: 10.3390/v17060828
Synergizing Attribute-Guided Latent Space Exploration (AGLSE) with Classical Molecular Simulations to Design Potent Pep-Magnet Peptide Inhibitors to Abrogate SARS-CoV-2 Host Cell Entry
Abstract
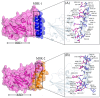
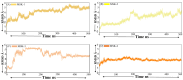
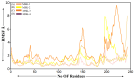
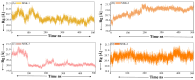
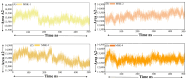
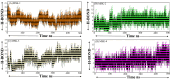
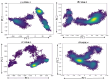
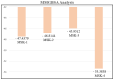
The COVID-19 infection, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has evoked a worldwide pandemic. Even though vaccines have been developed on an enormous scale, but due to regular mutations in the viral gene and the emergence of new strains could pose a more significant problem for the population. Therefore, new treatments are always necessary to combat future pandemics. Utilizing an antiviral peptide as a model biomolecule, we trained a generative deep learning algorithm on a database of known antiviral peptides to design novel peptide sequences with antiviral activity. Using artificial intelligence (AI), specifically variational autoencoders (VAE) and Wasserstein autoencoders (WAE), we were able to generate a latent space plot that can be surveyed for peptides with known properties and interpolated across a predictive vector between two defined points to identify novel peptides that exhibit dose-responsive antiviral activity. Two hundred peptide sequences were generated from the trained latent space and the top peptides were subjected to a molecular docking study. The docking analysis revealed that the top four peptides (MSK-1, MSK-2, MSK-3, and MSK-4) exhibited the strongest binding affinity, with docking scores of -106.4, -126.2, -125.7, and -127.8, respectively. Molecular dynamics simulations lasting 500 ns were performed to assess their stability and binding interactions. Further analyses, including MMGBSA, RMSD, RMSF, and hydrogen bond analysis, confirmed the stability and strong binding interactions of the peptide-protein complexes, suggesting that MSK-4 is a promising therapeutic agent for further development. We believe that the peptides generated through AI and MD simulations in the current study could be potential inhibitors in natural systems that can be utilized in designing therapeutic strategies against SARS-CoV-2.
Keywords: Omicron variant; SARS-CoV-2; Wasserstein autoencoders (WAE); deep learning; molecular docking; molecular dynamics simulation; variational autoencoders (VAE).
Conflict of interest statement
The author declares no conflict of interest.
Figures



Similar articles
-
Corticosteroid Prednisolone and flavonoid Chrysin as drug candidates against SARS-CoV-2 replication: Computational and experimental findings.Microb Pathog. 2025 Oct;207:107923. doi: 10.1016/j.micpath.2025.107923. Epub 2025 Jul 25. Microb Pathog. 2025. PMID: 40716470
-
Synthetic host defense peptide inhibits SARS-CoV-2 replication in vitro.Antimicrob Agents Chemother. 2025 Aug 6;69(8):e0170024. doi: 10.1128/aac.01700-24. Epub 2025 Jun 23. Antimicrob Agents Chemother. 2025. PMID: 40548715 Free PMC article.
-
A Rationally Designed Synthetic Antiviral Peptide Binder Targeting the Receptor-Binding Domain of SARS-CoV-2.J Phys Chem B. 2024 May 16;128(19):4631-4645. doi: 10.1021/acs.jpcb.4c00241. Epub 2024 Apr 24. J Phys Chem B. 2024. PMID: 38657271
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Samad A., Ajmal A., Mahmood A., Khurshid B., Li P., Jan S.M., Rehman A.U., He P., Abdalla A.N., Umair M., et al. Identification of novel inhibitors for SARS-CoV-2 as therapeutic options using machine learning-based virtual screening, molecular docking and MD simulation. Front. Mol. Biosci. 2023;10:1060076. doi: 10.3389/fmolb.2023.1060076. - DOI - PMC - PubMed
-
- Jarvis L.M. THE NEW DRUGS of 2019 the 48 medicines represent another highly productive year for the pharmaceutical industry, with cancer and rare-disease drugs again dominating the list. Chem. Eng. News. 2020;98:30–36.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

