Integrative Computational Approaches for Understanding Drug Resistance in HIV-1 Protease Subtype C
- PMID: 40573441
- PMCID: PMC12197794
- DOI: 10.3390/v17060850
Integrative Computational Approaches for Understanding Drug Resistance in HIV-1 Protease Subtype C
Abstract
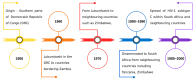
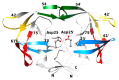
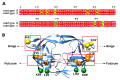
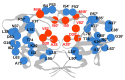
Acquired immunodeficiency syndrome (AIDS) is a chronic disease condition caused by the human immunodeficiency virus (HIV). The widespread availability of highly active antiretroviral therapies has helped to control HIV. There are ten FDA-approved protease inhibitors (PIs) that are used as part of antiretroviral therapies in HIV treatment. Importantly, all these drugs are designed and developed against the protease (PR) from HIV subtype B. On the other hand, HIV-1 PR subtype C, which is the most dominant strain in countries including South Africa and India, has shown resistance to PIs due to its genetic diversity and varied mutations. The emergence of resistance is concerning because the virus continues to replicate despite treatment; hence, it is necessary to develop drugs specifically against subtype C. This review focuses on the origin, genetic diversity, and mutations associated with HIV-1 PR subtype C. Furthermore, computational studies performed on HIV-1 PR subtype C and mutations associated with its resistance to PIs are highlighted. Moreover, potential research gaps and future directions in the study of HIV-1 PR subtype C are discussed.
Keywords: AIDS; HIV protease; MD simulations; computational studies; insertion; mutation; structure activity relationship; subtype C.
Conflict of interest statement
We state that there is no conflict of interest.
Figures
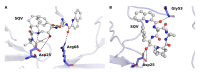
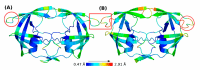
Similar articles
-
Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: a systematic review of virologic and biochemical evidence.AIDS Rev. 2008 Oct-Dec;10(4):212-23. AIDS Rev. 2008. PMID: 19092977
-
Contrasting the effect of hinge region insertions and non-active site mutations on HIV protease-inhibitor interactions: Insights from altered flap dynamics.J Mol Graph Model. 2024 Dec;133:108850. doi: 10.1016/j.jmgm.2024.108850. Epub 2024 Aug 29. J Mol Graph Model. 2024. PMID: 39226791
-
Structural Implications of HIV-1 Protease Subtype C Bound to Darunavir: A Molecular Dynamics Study.Proteins. 2025 Sep;93(9):1426-1435. doi: 10.1002/prot.26817. Epub 2025 Mar 3. Proteins. 2025. PMID: 40026243
-
Trends in HIV-1 pretreatment drug resistance and HIV-1 variant dynamics among antiretroviral therapy-naive Ethiopians from 2003 to 2018: a pooled sequence analysis.Virol J. 2023 Oct 25;20(1):243. doi: 10.1186/s12985-023-02205-w. Virol J. 2023. PMID: 37880705 Free PMC article.
-
Plasma Viral Load of 200 Copies/mL is a Suitable Threshold to Define Viral Suppression and HIV Drug Resistance Testing in Low- and Middle-Income Countries: Evidence From a Facility-Based Study in Cameroon.J Int Assoc Provid AIDS Care. 2024 Jan-Dec;23:23259582241306484. doi: 10.1177/23259582241306484. J Int Assoc Provid AIDS Care. 2024. PMID: 39711049 Free PMC article.
References
-
- Barré-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vézinet-Brun F., Rouzioux C., et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. - DOI - PubMed
-
- UNAIDS Global HIV & AIDS Statistics—Fact Sheet. [(accessed on 20 February 2025)]. Available online: https://www.unaids.org/en/resources/fact-sheet.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

