Discovery of a Novel Parahenipavirus, Parahenipavirus_GH, in Shrews in South Korea, 2022
- PMID: 40573458
- PMCID: PMC12197346
- DOI: 10.3390/v17060867
Discovery of a Novel Parahenipavirus, Parahenipavirus_GH, in Shrews in South Korea, 2022
Abstract
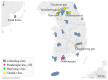
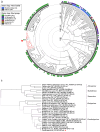
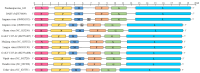
Highly pathogenic henipaviruses (Nipah and Hendra viruses) and parahenipaviruses (Langya virus) have demonstrated significant zoonotic potential. We aimed to identify Henipavirus or Parahenipavirus species in rodents and shrews in South Korea to underline the potential zoonotic transmission risk. Kidney and lung tissues from 285 rodents and shrews were screened for Henipavirus and Parahenipavirus using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) targeting the Gamak virus and Daeryong virus (DARV) sequences. Based on the qRT-PCR results, 75 out of the 285 individuals tested positive, with the highest viral loads in the kidneys of Apodemus agrarius, Crocidura lasiura, and Crocidura shantungensis. A kidney sample from C. shantungensis that exhibited the lowest Ct value was further analyzed using PCR, Sanger sequencing, and metagenomic analysis, yielding a near-complete genome of a novel Parahenipavirus, designated Parahenipavirus_GH (PHNV-GH), which is phylogenetically related to DARV and Jingmen virus but exhibits distinct genomic features. Ixodes granulatus ticks were also identified on the host shrew. The identification of PHNV-GH in southern South Korea expands the known geographical distribution range of parahenipaviruses and highlights the ongoing risk of zoonotic transmission. Given the uncertain transmission dynamics and pathogenic potential of parahenipaviruses, comprehensive environmental surveillance and characterization of emerging parahenipaviruses are essential for preventing future outbreaks.
Keywords: Henipavirus; Parahenipavirus; Paramyxoviridae; shrew; zoonotic.
Conflict of interest statement
The authors declare that they have no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Trafficking and Activation of Henipavirus, Parahenipavirus, and Henipa-like Virus Fusion Proteins.Viruses. 2025 Jun 19;17(6):866. doi: 10.3390/v17060866. Viruses. 2025. PMID: 40573457 Free PMC article. Review.
-
Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea.Viruses. 2021 Oct 7;13(10):2020. doi: 10.3390/v13102020. Viruses. 2021. PMID: 34696450 Free PMC article.
-
Infectome analysis of bat kidneys from Yunnan province, China, reveals novel henipaviruses related to Hendra and Nipah viruses and prevalent bacterial and eukaryotic microbes.PLoS Pathog. 2025 Jun 24;21(6):e1013235. doi: 10.1371/journal.ppat.1013235. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40554741 Free PMC article.
-
Discovery and Genomic Characterization of a Novel Henipavirus, Angavokely Virus, from Fruit Bats in Madagascar.J Virol. 2022 Sep 28;96(18):e0092122. doi: 10.1128/jvi.00921-22. Epub 2022 Aug 30. J Virol. 2022. PMID: 36040175 Free PMC article.
-
Zoonotic Paramyxoviruses: Evolution, Ecology, and Public Health Strategies in a Changing World.Viruses. 2024 Oct 29;16(11):1688. doi: 10.3390/v16111688. Viruses. 2024. Retraction in: Viruses. 2025 Jul 16;17(7):992. doi: 10.3390/v17070992. PMID: 39599803 Free PMC article. Retracted. Review.
References
-
- Genus: Henipavirus. [(accessed on 18 August 2023)]. Available online: https://ictv.global/report/chapter/paramyxoviridae/paramyxoviridae/henip....
-
- Taxon Name: Parahenipavirus. [(accessed on 29 May 2025)]. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202415744&taxon_nam....
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

