Advances in Viroporin Function and Structure: A Comparative Analysis of Alphavirus 6K with Well-Characterized Viroporins
- PMID: 40573459
- PMCID: PMC12197650
- DOI: 10.3390/v17060868
Advances in Viroporin Function and Structure: A Comparative Analysis of Alphavirus 6K with Well-Characterized Viroporins
Abstract
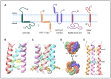
Viruses encode ion channel proteins called viroporins to assist in infection and immune evasion. The alphavirus 6K protein is classified as a member of the viroporin family of proteins. Several studies have characterized the role of 6K in alphavirus budding and infection since its discovery in the late 1970s. In this review, we summarize 6K research and discuss some unanswered questions regarding 6K biology. We highlight the similarities and differences between 6K and viroporins of clinically relevant viruses-influenza A virus, HIV-1, hepatitis C virus, and SARS-CoV-2-and address their importance as therapeutic targets. The sensitivity of these viroporins to common inhibitors and their ability to functionally complement each other underscore their potential as targets for broad-spectrum antiviral therapies.
Keywords: HCV p7; HIV-1 Vpu; IAV M2; SARS-CoV-2 envelope; alphavirus 6K; alphavirus TF; antivirals; viroporins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Antiviral effect of the viroporin inhibitors against Taiwan isolates of infectious bronchitis virus (IBV).Virus Res. 2024 Nov;349:199458. doi: 10.1016/j.virusres.2024.199458. Epub 2024 Aug 27. Virus Res. 2024. PMID: 39187047 Free PMC article.
-
Viroporins beyond pathogenesis: Ion channel properties as the key to unlocking a neglected antiviral target.Pharmacol Res. 2025 Sep;219:107863. doi: 10.1016/j.phrs.2025.107863. Epub 2025 Jul 17. Pharmacol Res. 2025. PMID: 40683481 Review.
-
Advanced Computational Approaches to Evaluate the Potential of New-Generation Adamantane-Based Drugs as Viroporin Inhibitors: A Case Study on SARS-CoV-2.J Phys Chem B. 2025 Aug 14;129(32):8127-8143. doi: 10.1021/acs.jpcb.5c02898. Epub 2025 Aug 4. J Phys Chem B. 2025. PMID: 40758312
-
The envelope proteins from SARS-CoV-2 and SARS-CoV potently reduce the infectivity of human immunodeficiency virus type 1 (HIV-1).Retrovirology. 2022 Nov 19;19(1):25. doi: 10.1186/s12977-022-00611-6. Retrovirology. 2022. PMID: 36403071 Free PMC article.
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

