Comparative Transcriptome and Metabolome Profiling Revealed Molecular Cascade Events During the Enzymatic Browning of Potato Tubers After Cutting
- PMID: 40573805
- PMCID: PMC12196699
- DOI: 10.3390/plants14121817
Comparative Transcriptome and Metabolome Profiling Revealed Molecular Cascade Events During the Enzymatic Browning of Potato Tubers After Cutting
Abstract
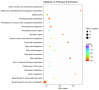
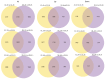
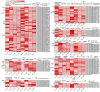
Enzymatic browning is a major issue in potato processing, causing a decline in both nutritional value and quality. Although there are numerous studies on the mechanism of enzymatic browning of potato tubers, few relevant reports are available on the changes at the transcriptome level during enzymatic browning as well as on the differences in the browning process of potato tubers with differing degrees of enzymatic browning potential. To gain insights into the molecular mechanism of enzymatic browning after cutting, this study presents the transcriptional characterization of temporal molecular events during enzymatic browning of browning-resistant (BR) and browning-susceptible (BS) potato tubers. RNA-sequencing (RNA-seq) analysis detected 19,377 and 13,741 differentially expressed genes (DEGs) in BR and BS tubers, respectively, with similar function enrichment observed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Up-regulated DEGs were significantly enriched in the pathways related to phenol and lipid biosynthesis, while the down-regulated DEGs were significantly enriched in the pathways related to programmed cell death. Significant redox-related pathways occurred earlier in BS tubers compared to the BR tubers. Further analysis revealed that the BS tubers had a stronger phenolic synthesis ability compared to the BR tubers. However, the BR tubers showed a stronger free radical scavenging ability compared to the BS tubers. The results of our study provide insights into the temporal molecular events that occur during the enzymatic browning of potato tubers after cutting and the potential molecular mechanisms for different degrees of enzymatic browning.
Keywords: cut-wounding; enzymatic browning; metabolome; potato tubers; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
Systematic review and economic analysis of the comparative effectiveness of different inhaled corticosteroids and their usage with long-acting beta2 agonists for the treatment of chronic asthma in adults and children aged 12 years and over.Health Technol Assess. 2008 May;12(19):iii-iv, 1-360. doi: 10.3310/hta12190. Health Technol Assess. 2008. PMID: 18485271
-
Insights into the molecular mechanisms of browning tolerance in luffa: a transcriptome and metabolome analysis.Front Plant Sci. 2025 Jun 10;16:1530531. doi: 10.3389/fpls.2025.1530531. eCollection 2025. Front Plant Sci. 2025. PMID: 40556689 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Integrated Transcriptome and Metabolome Analysis Provides Insights into the Low-Temperature Response in Sweet Potato (Ipomoea batatas L.).Genes (Basel). 2025 Jul 28;16(8):899. doi: 10.3390/genes16080899. Genes (Basel). 2025. PMID: 40869948 Free PMC article.
-
Physical exercise training interventions for children and young adults during and after treatment for childhood cancer.Cochrane Database Syst Rev. 2016 Mar 31;3(3):CD008796. doi: 10.1002/14651858.CD008796.pub3. Cochrane Database Syst Rev. 2016. PMID: 27030386 Free PMC article.
References
-
- Walker J.R. Enzymatic browning in fruits: Its biochemistry and control. In: Lee C.Y., Whitaker J.R., editors. Enzymatic Browning and Its Prevention. 1st ed. Volume 600. ACS Publications; Washington, DC, USA: 1995. pp. 8–22.
-
- He Q., Luo Y. Enzymatic browning and its control in fresh-cut produce. Stewart Postharvest Rev. 2007;6:3. doi: 10.2212/spr.2007.6.3. - DOI
-
- Richard-Forget F.C., Gauillard F.A. Oxidation of chlorogenic acid, catechins, and 4-Methylcatechol in model solutions by combinations of pear (Pyrus communis Cv. Williams) polyphenol oxidase and peroxidase: A Possible involvement of peroxidase in enzymatic browning. J. Agric. Food Chem. 1997;45:2472–2476. doi: 10.1021/jf970042f. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

