A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation
- PMID: 40573810
- PMCID: PMC12196923
- DOI: 10.3390/plants14121821
A Non-Specific Phytohormone Regulatory Network in Saccharina japonica Coordinates Growth and Environmental Adaptation
Abstract
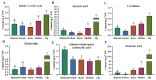
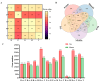
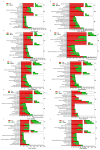
Saccharina japonica (S. japonica) is a large-scale intertidal aquatic plant that exhibits characteristics such as rhizoid, holdfast, and blade differentiation. It demonstrates remarkable environmental adaptability. However, compared with higher plants, details about its phytohormone content, distribution, synthesis, and accumulation remain poorly understood. In this study, the phytohormone contents distribution and expression patterns of synthetic genes in different parts of S. japonica, including the rhizoid, petiole, basis, middle, and tip, were analyzed in detail by combining targeted metabolomics and transcriptomics analyses. A total of 20 phytohormones were detected in S. japonica, including auxin, abscisic acid (ABA), cytokinin (CTK), ethylene (ETH), gibberellin (GA), jasmonate acid (JA), and salicylic acid (SA), with significant site-differentiated accumulation. ABA and JA were significantly enriched in the tips (28.01 ng·g-1 FW and 170.67 ng·g-1 FW, respectively), whereas SA accumulated specifically only in the rhizoid. We also identified 12 phytohormones, such as gibberellin A1, methyl jasmonate, and trans-zeatin for the first time in S. japonica. Transcriptomic profiling revealed the tissue-specific expression of phytohormone biosynthesis genes, such as CYP735A (CTK synthesis), in the rhizoids and LOX/NCED (JA/ABA synthesis) in the tips. Key pathways, such as carotenoid biosynthesis and cysteine methionine metabolism, were found to be differentially enriched across tissues, aligning with hormone accumulation patterns. Additionally, an enrichment analysis of differentially expressed genes between various parts indicated that different parts of S. japonica performed distinct functions even though it does not have organ differentiation. This study is the first to uncover the distribution characteristics of phytohormones and their synthetic differences in different parts of S. japonica and elucidates how S. japonica achieves functional specialization through non-specific phytohormone regulation despite lacking organ differentiation, which provides an important theoretical basis for research on the developmental biology of macroalgae and their mechanisms of response to adversity.
Keywords: Saccharina japonica; abiotic stress; metabolomics; phytohormones; tissue-specific adaptation; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Unveiling the secrets of abiotic stress tolerance in plants through molecular and hormonal insights.3 Biotech. 2024 Oct;14(10):252. doi: 10.1007/s13205-024-04083-7. Epub 2024 Sep 26. 3 Biotech. 2024. PMID: 39345964 Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
From Hormones to Harvests: A Pathway to Strengthening Plant Resilience for Achieving Sustainable Development Goals.Plants (Basel). 2025 Jul 27;14(15):2322. doi: 10.3390/plants14152322. Plants (Basel). 2025. PMID: 40805671 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
References
-
- Meng X. The Progress of the Methods to Identify and Classify Phytoplankton. Biol. Chem. Eng. 2019;5:102–104.
-
- Li X., Zhong C., Jin Z., Lin Q., Shan T., Pang S. Genetic structure, major chemical component contents and agronomic characters of the principal cultivars of the kelp Saccharina japonica in the largest kelp production province in China. J. Appl. Phycol. 2023;35:763–772. doi: 10.1007/s10811-023-02905-4. - DOI
-
- Gao G., Beardall J., Jin P., Gao L., Xie S., Gao K. A review of existing and potential blue carbon contributions to climate change mitigation in the Anthropocene. J. Appl. Ecol. 2022;59:1686–1699. doi: 10.1111/1365-2664.14173. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

